Full Text |
QUERY: Citation Author = "Rubinstein, J.L." | MyPDB Login | Search API |
| Search Summary | This query matches 237 Structures. |
Structure Determination MethodologyScientific Name of Source OrganismMore... TaxonomyExperimental MethodPolymer Entity TypeRefinement Resolution (Å)Release DateEnzyme Classification NameMembrane Protein AnnotationSymmetry TypeSCOP Classification | 1 to 25 of 237 Structures Page 1 of 10 Sort by
Solution structure of Bacteriophage Lambda gpFIIMaxwell, K.L., Cardarelli, L., Neudecker, P., Davidson, A.R., Ontario Centre for Structural Proteomics (OCSP) (2010) Proc Natl Acad Sci U S A 107: 14384-14389
NMR solution structure of peptide a2N(1-17) from Mus musculus V-ATPaseDip, P., Gruber, G., Marshansky, V. (2013) J Biological Chem 288: 5896-5913
Structure of FepE- Bacterial Polysaccharide Co-polymeraseTocilj, A., Matte, A., Cygler, M. (2008) Nat Struct Mol Biol 15: 130-138
Structure of FepE- Bacterial Polysaccharide Co-polymeraseTocilj, A., Matte, A., Cygler, M. (2008) Nat Struct Mol Biol 15: 130-138
Structure of WzzE- Bacterial Polysaccharide Co-polymeraseTocilj, A., Matte, A., Cygler, M. (2008) Nat Struct Mol Biol 15: 130-138
Fragment of WzzB, Polysaccharide Co-polymerase from Salmonella TyphimuriumTocilj, A., Matte, A., Cygler, M. (2008) Nat Struct Mol Biol 15: 130-138
Fitted atomic models of Thermus thermophilus V-ATPase subunits into cryo-EM map(2012) Nature 481: 214-218
Yeast V-ATPase state 1Zhao, J., Benlekbir, S., Rubinstein, J.L. (2015) Nature 521: 241-245
Yeast V-ATPase state 2Zhao, J., Benlekbir, S., Rubinstein, J.L. (2015) Nature 521: 241-245
Yeast V-ATPase state 3Zhao, J., Benlekbir, S., Rubinstein, J.L. (2015) Nature 521: 241-245
Crystal structure of bacteriophage HK97 gp6Lam, R., Tuite, A., Battaile, K.P., Edwards, A.M., Maxwell, K.L., Chirgadze, N.Y. (2010) J Mol Biology 395: 754-768
Crystal structure of the SPOP BTB domain complexed with the Cul3 N-terminal domain(2012) Structure 20: 1141-1153
Crystal structure of full-length mouse alphaE-catenin(2013) J Biological Chem 288: 15913-15925
Crystal structure of the alphaN-catenin actin-binding domain(2013) J Biological Chem 288: 15913-15925
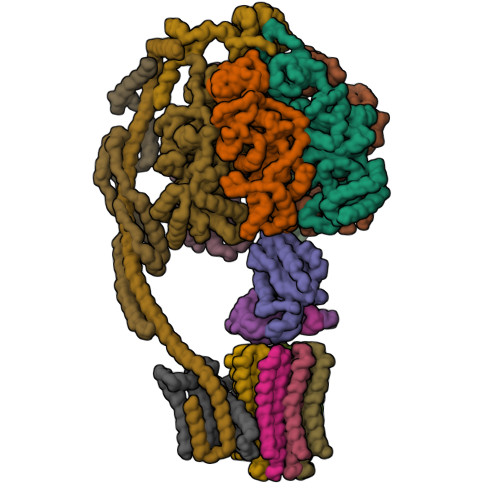
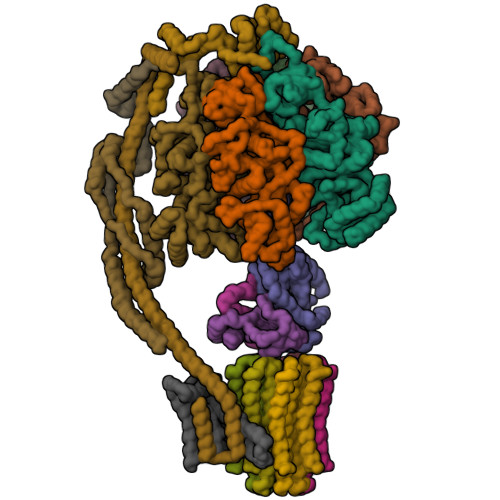
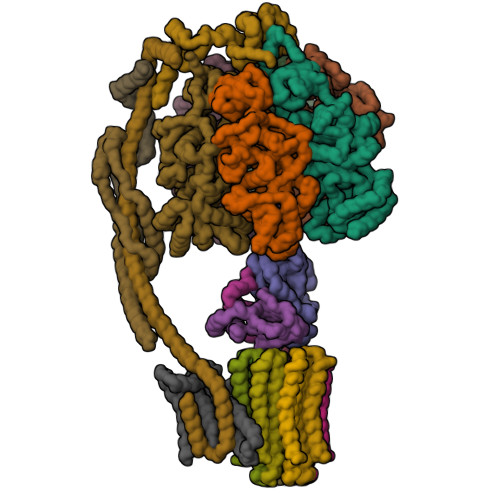
Bovine mitochondrial ATP synthase state 1aZhou, A., Rohou, A., Schep, D.G., Bason, J.V., Montgomery, M.G., Walker, J.E., Grigorieff, N., Rubinstein, J.L. (2015) Elife 4: e10180-e10180
Bovine mitochondrial ATP synthase state 1bZhou, A., Rohou, A., Schep, D.G., Bason, J.V., Montgomery, M.G., Walker, J.E., Grigorieff, N., Rubinstein, J.L. (2015) Elife 4: e10180-e10180
Bovine mitochondrial ATP synthase state 2aZhou, A., Rohou, A., Schep, D.G., Bason, J.V., Montgomery, M.G., Walker, J.E., Grigorieff, N., Rubinstein, J.L. (2015) Elife 4: e10180-e10180
Bovine mitochondrial ATP synthase state 2bZhou, A., Rohou, A., Schep, D.G., Bason, J.V., Montgomery, M.G., Walker, J.E., Grigorieff, N., Rubinstein, J.L. (2015) Elife 4: e10180-e10180
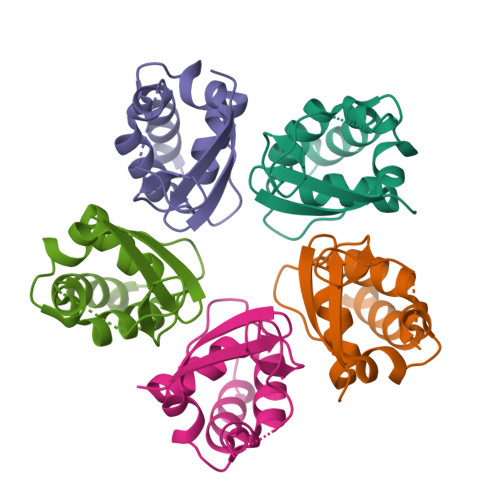
Crystal structure of pentameric KCTD1 BTB domain form 1Ji, A.X., Chu, A., Prive, G.G. (2016) J Mol Biology 428: 92-107
Crystal structure of pentameric KCTD1 BTB domain form 2Ji, A.X., Chu, A., Prive, G.G. (2016) J Mol Biology 428: 92-107
Crystal structure of pentameric KCTD9 BTB domainJi, A.X., Chu, A., Prive, G.G. (2016) J Mol Biology 428: 92-107
Bovine mitochondrial ATP synthase state 2cZhou, A., Rohou, A., Schep, D.G., Bason, J.V., Montgomery, M.G., Walker, J.E., Grigorieff, N., Rubinstein, J.L. (2015) Elife 4: e10180-e10180
Bovine mitochondrial ATP synthase state 3aZhou, A., Rohou, A., Schep, D.G., Bason, J.V., Montgomery, M.G., Walker, J.E., Grigorieff, N., Rubinstein, J.L. (2015) Elife 4: e10180-e10180
Bovine mitochondrial ATP synthase state 3bZhou, A., Rohou, A., Schep, D.G., Bason, J.V., Montgomery, M.G., Walker, J.E., Grigorieff, N., Rubinstein, J.L. (2015) Elife 4: e10180-e10180
Structure of the ADP-bound VAT complexHuang, R., Ripstein, Z.A., Augustyniak, R., Lazniewski, M., Ginalski, K., Kay, L.E., Rubinstein, J.L. (2016) Proc Natl Acad Sci U S A 113: E4190
1 to 25 of 237 Structures Page 1 of 10 Sort by |