Full Text |
QUERY: Citation Author = "Taneja, A." | MyPDB Login | Search API |
| Search Summary | This query matches 67 Structures. |
Structure Determination MethodologyScientific Name of Source OrganismMore... TaxonomyExperimental MethodPolymer Entity TypeRefinement Resolution (Å)Release DateEnzyme Classification NameMembrane Protein AnnotationSymmetry TypeSCOP Classification | 1 to 25 of 67 Structures Page 1 of 3 Sort by
Structure of succinyl-CoA: 3-ketoacid CoA transferase from Drosophila melanogasterWang, Y.C., Shi, Z.B., Zhang, M. (2013) Acta Crystallogr Sect F Struct Biol Cryst Commun 69: 1089-1093
Crystal structure of zebrafish MO25Zhang, Z.Z., Shi, Z.B., Zhang, M. (2013) Acta Crystallogr Sect F Struct Biol Cryst Commun 69: 989-993
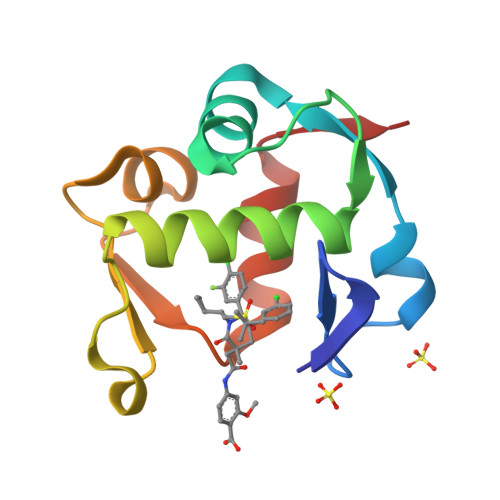
co-crystal structure of MDM2 (17-111) in complex with compound 25(2014) J Med Chem 57: 1454-1472
Co-crystal Structure of MDM2 in Complex with AM-7209Shaffer, P.L., Huang, X., Yakowec, P., Long, A.M. (2014) J Med Chem 57: 10499-10511
Misting with CDAChin, K.H., Chen, C.K., Tu, Z.I., Chou, S.H. (2015) Biochemistry 54: 4936-4951
Structure of RCK domain with cda(2015) Biochemistry 54: 4936-4951
Crystal structure of Phytophthora. sojae PSR2(2019) Proc Natl Acad Sci U S A
Structure of MshE with cdg(2016) Nat Commun 7: 12481-12481
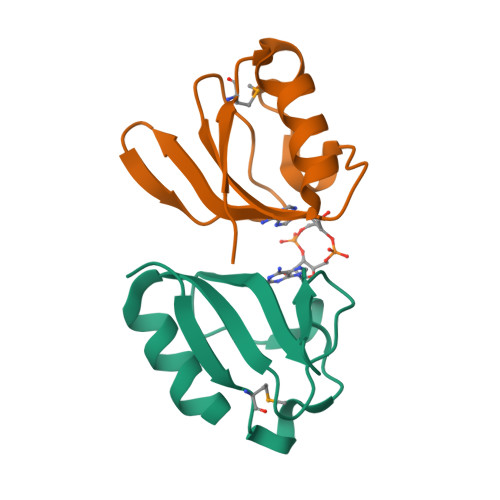
Crystal structure of NlpC/p60 domain of peptidoglycan hydrolase SagAKim, B., Oren, D.A., Hang, H.C. (2019) Elife 8:
SARS-CoV-2 main protease (Mpro) apo structure (space group P212121)Wang, Y.C., Yang, C.S., Hou, M.H., Tsai, C.L., Chou, Y.Z., Chen, Y. (2020) Am J Cancer Res 10: 2535-2545
1.7A resolution structure of SARS-CoV-2 main protease (Mpro) in complex with broad-spectrum coronavirus protease inhibitor GC376Wang, Y.C., Yang, C.S., Hou, M.H., Tsai, C.L., Chou, Y.Z., Chen, Y., Hung, M.C. (2020) Am J Cancer Res 10: 2535-2545
apo-form cyclic trinucleotide synthase CdnDYang, C.-S., Hou, M.-H., Tsai, C.-L., Wang, Y.-C., Ko, T.-P., Chen, Y. (2021) Nucleic Acids Res 49: 4725-4737
ddATP complex of cyclic trinucleotide synthase CdnDYang, C.-S., Hou, M.-H., Tsai, C.-L., Wang, Y.-C., Ko, T.-P., Chen, Y. (2021) Nucleic Acids Res 49: 4725-4737
cyclic trinucleotide synthase CdnD in complex with ATP and ADPYang, C.-S., Hou, M.-H., Tsai, C.-L., Wang, Y.-C., Ko, T.-P., Chen, Y. (2021) Nucleic Acids Res 49: 4725-4737
apo-form cyclic trinucleotide synthase CdnDYang, C.-S., Hou, M.-H., Tsai, C.-L., Wang, Y.-C., Ko, T.-P., Chen, Y. (2021) Nucleic Acids Res 49: 4725-4737
ATP complex with double mutant cyclic trinucleotide synthase CdnDYang, C.-S., Hou, M.-H., Tsai, C.-L., Wang, Y.-C., Ko, T.-P., Chen, Y. (2021) Nucleic Acids Res 49: 4725-4737
Crystal structure of the N-terminal domain of TagH from Bacillus subtilisKo, T.P., Yang, C.S., Wang, Y.C., Chen, Y. (2020) Biochem Biophys Res Commun 536: 1-6
Crystal structure of SARS-CoV-2 main protease in complex with TafenoquineChen, Y., Wang, Y.C., Yang, C.S., Hung, M.C. (2022) J Biological Chem 298: 101658-101658
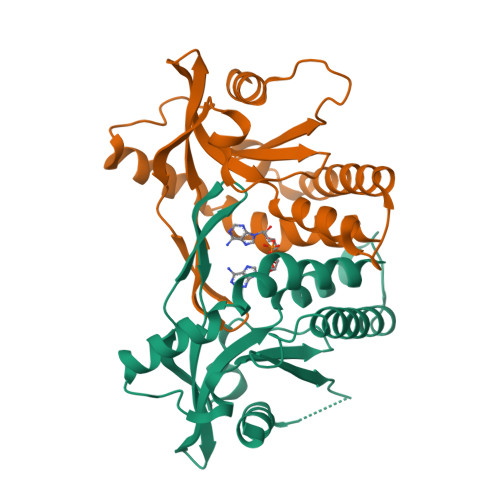
Complex Structure of antibody BD-503 and RBD of COVID-19Xu, H., Wang, B., Zhao, T.N., Su, X.D. (2021) Cell Res 31: 1126-1129
Complex Structure of antibody BD-503 and RBD-S477N of COVID-19Xu, H., Wang, B., Zhao, T.N., Su, X.D. (2021) Cell Res 31: 1126-1129
Complex Structure of antibody BD-503 and RBD-N501Y of COVID-19Xu, H., Wang, B., Zhao, T.N., Su, X.D. (2021) Cell Res 31: 1126-1129
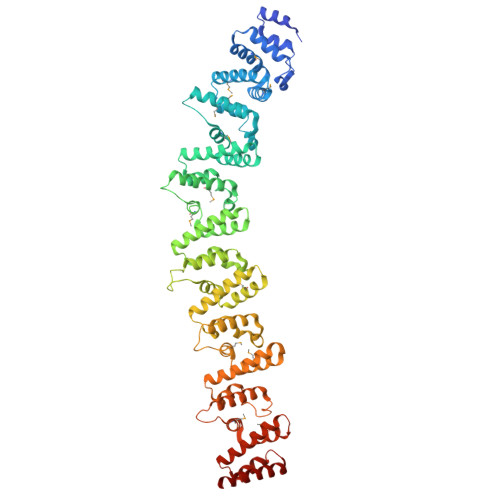
Structure and Activity of SLAC1 Channels for Stomatal Signaling in LeavesDeng, Y., Kashtoh, H., Wang, Q., Zhen, G., Li, Q., Tang, L., Gao, H., Zhang, C., Qin, L., Su, M., Li, F., Huang, X., Wang, Y., Xie, Q., Clarke, O.B., Hendrickson, W.A., Chen, Y. (2021) Proc Natl Acad Sci U S A 118:
Complex Structure of antibody BD-503 and RBD-E484K of COVID-19Xu, H., Wang, B., Zhao, T.N., Su, X.D. (2021) Cell Res 31: 1126-1129
Complex Structure of antibody BD-503 and RBD-501Y.V2 of COVID-19Xu, H., Wang, B., Zhao, T.N., Su, X.D. (2021) Cell Res 31: 1126-1129
Crystal structure of SARS-CoV-2 main protease in complex with Z-DEVD-FMKWang, Y.C., Yang, C.S., Hou, M.H., Tsai, C.L., Chen, Y. To be published
1 to 25 of 67 Structures Page 1 of 3 Sort by |