Full Text |
QUERY: Chemical ID(s) = HZP | MyPDB Login | Search API |
| Search Summary | This query matches 6 Polymer Entities. |
Structure Determination MethodologyScientific Name of Source OrganismTaxonomyExperimental MethodPolymer Entity TypeRefinement Resolution (Å)Release DateEnzyme Classification NameSymmetry TypeSCOP Classification | 1 to 6 of 6 Polymer Entities Page 1 of 1 Sort by
Crystal structure of the bacterial lysozyme from Streptomyces coelicolor at 1.65 A resolutionRau, A., Hogg, T., Marquardt, R., Hilgenfeld, R. (2001) J Biological Chem 276: 31994-31999
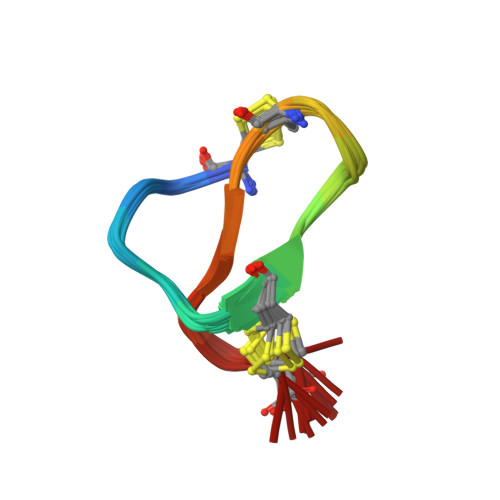
Structure of Sviceucin, an antibacterial type I lasso peptide from Streptomyces sviceusLi, Y., Ducasse, R., Blond, A., Zirah, S., Goulard, C., Lescop, E., Guittet, E., Pernodet, J., Rebuffat, S. To be published
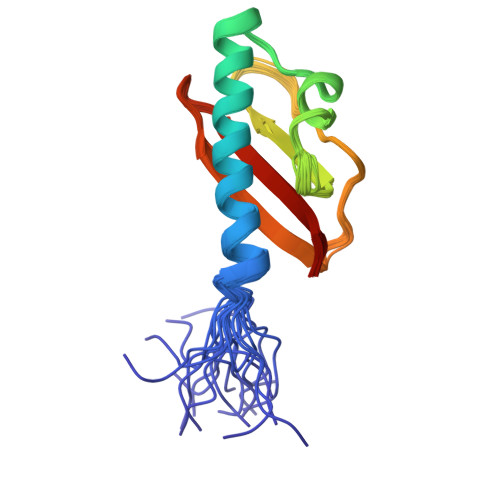
Solution structure of the extracellular sensor domain of DraK histidine kinase(2014) PLoS One 9: e107168-e107168
NMR solution structure of class IV lasso peptide felipeptin A1 from Amycolatopsis sp. YIM10Madland, E., Guerrero-Garzon, J.F., Zotchev, S.B., Aachmann, F.L., Courtade, G. (2020) iScience 23: 101785-101785
NMR solution structure of class IV lasso peptide felipeptin A2 from Amycolatopsis sp. YIM10Madland, E., Aachmann, F.L., Guerrero-Garzon, J.F., Zotchev, S.B., Courtade, G. (2020) iScience 23: 101785-101785
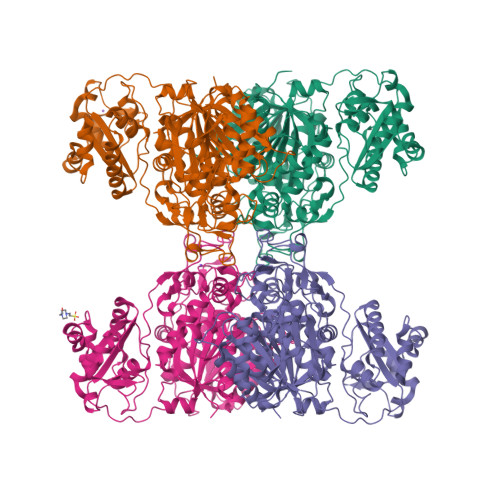
Structure of SalC, a gamma-lactam-beta-lactone bicyclase for salinosporamide biosynthesisChen, P.Y.-T., Trivella, D.B.B., Bauman, K.D., Moore, B.S. (2022) Nat Chem Biol 18: 538-546
1 to 6 of 6 Polymer Entities Page 1 of 1 Sort by |