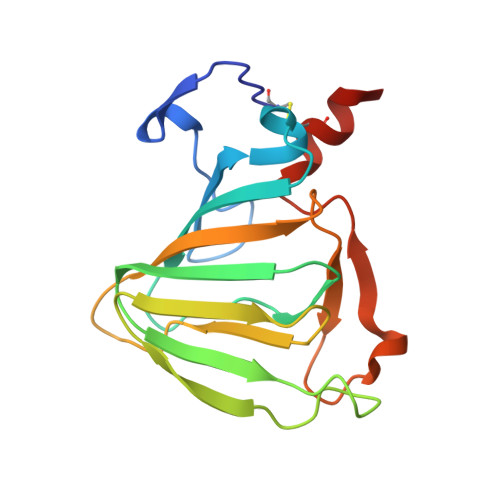
Crystal structure of auxin-binding protein 1 in complex with auxin.
Woo, E.J., Marshall, J., Bauly, J., Chen, J.G., Venis, M., Napier, R.M., Pickersgill, R.W.(2002) EMBO J 21: 2877-2885
- PubMed: 12065401
- DOI: https://doi.org/10.1093/emboj/cdf291
- Primary Citation of Related Structures:
1LR5, 1LRH - PubMed Abstract:
The structure of auxin-binding protein 1 (ABP1) from maize has been determined at 1.9 A resolution, revealing its auxin-binding site. The structure confirms that ABP1 belongs to the ancient and functionally diverse germin/seed storage 7S protein superfamily. The binding pocket of ABP1 is predominantly hydrophobic with a metal ion deep inside the pocket coordinated by three histidines and a glutamate. Auxin binds within this pocket, with its carboxylate binding the zinc and its aromatic ring binding hydrophobic residues including Trp151. There is a single disulfide between Cys2 and Cys155. No conformational rearrangement of ABP1 was observed when auxin bound to the protein in the crystal, but examination of the structure reveals a possible mechanism of signal transduction.
Organizational Affiliation:
Biological Sciences, Queen Mary, University of London, Mile End Road, London E1 4NS, UK.