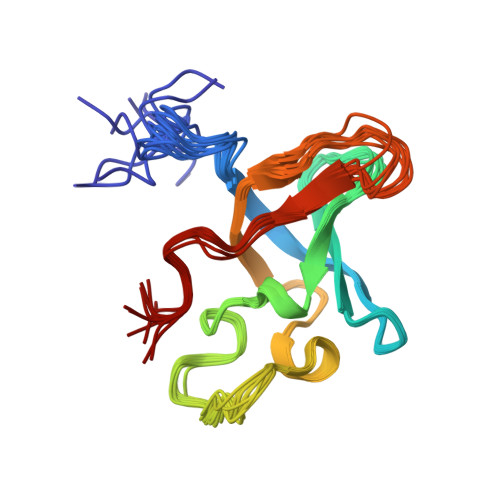
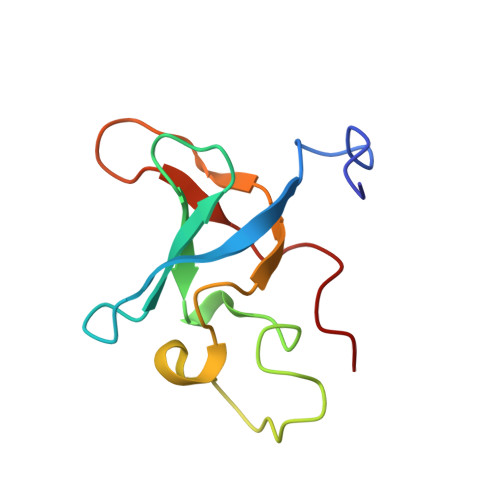
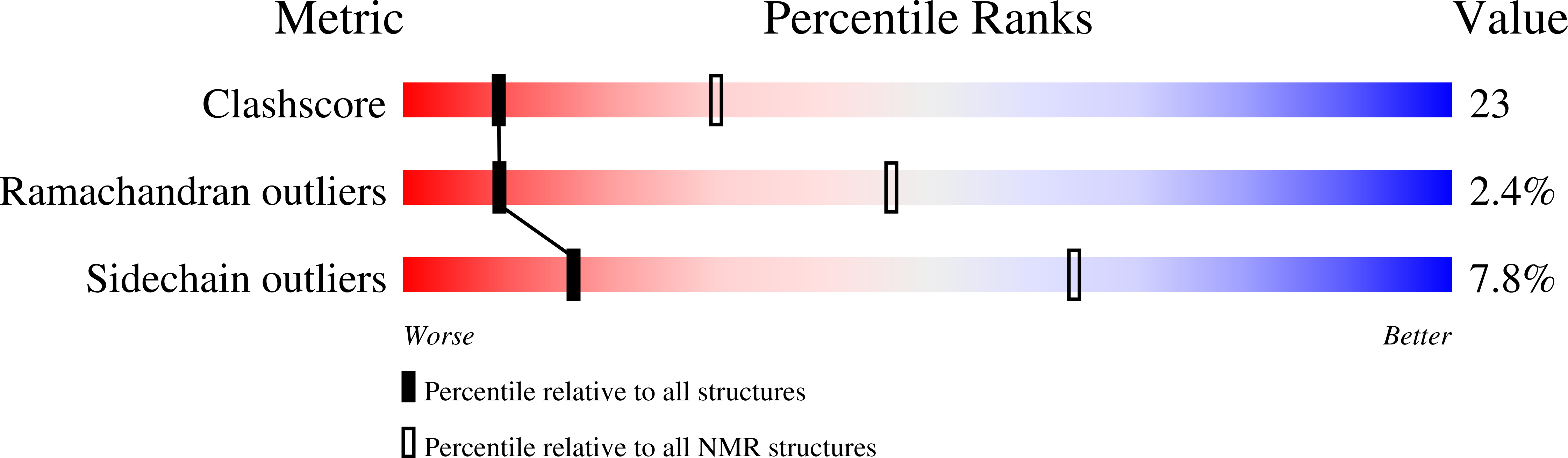Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces.
Schubert, M., Edge, R.E., Lario, P., Cook, M.A., Strynadka, N.C., Mackie, G.A., McIntosh, L.P.(2004) J Mol Biology 341: 37-54
- PubMed: 15312761
- DOI: https://doi.org/10.1016/j.jmb.2004.05.061
- Primary Citation of Related Structures:
1SLJ, 1SMX, 1SN8 - PubMed Abstract:
S1 domains occur in four of the major enzymes of mRNA decay in Escherichia coli: RNase E, PNPase, RNase II, and RNase G. Here, we report the structure of the S1 domain of RNase E, determined by both X-ray crystallography and NMR spectroscopy. The RNase E S1 domain adopts an OB-fold, very similar to that found with PNPase and the major cold shock proteins, in which flexible loops are appended to a well-ordered five-stranded beta-barrel core. Within the crystal lattice, the protein forms a dimer stabilized primarily by intermolecular hydrophobic packing. Consistent with this observation, light-scattering, chemical crosslinking, and NMR spectroscopic measurements confirm that the isolated RNase E S1 domain undergoes a specific monomer-dimer equilibrium in solution with a K(D) value in the millimolar range. The substitution of glycine 66 with serine dramatically destabilizes the folded structure of this domain, thereby providing an explanation for the temperature-sensitive phenotype associated with this mutation in full-length RNase E. Based on amide chemical shift perturbation mapping, the binding surface for a single-stranded DNA dodecamer (K(D)=160(+/-40)microM) was identified as a groove of positive electrostatic potential containing several exposed aromatic side-chains. This surface, which corresponds to the conserved ligand-binding cleft found in numerous OB-fold proteins, lies distal to the dimerization interface, such that two independent oligonucleotide-binding sites can exist in the dimeric form of the RNase E S1 domain. Based on these data, we propose that the S1 domain serves a dual role of dimerization to aid in the formation of the tetrameric quaternary structure of RNase E as described by Callaghan et al. in 2003 and of substrate binding to facilitate RNA hydrolysis by the adjacent catalytic domains within this multimeric enzyme.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, The University of British Columbia, Vancouver, BC, Canada V6T 1Z3.