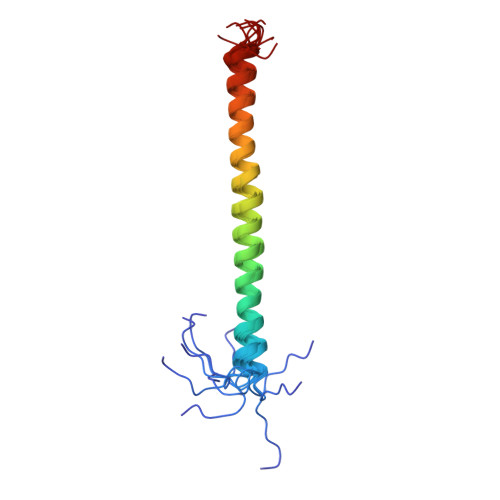
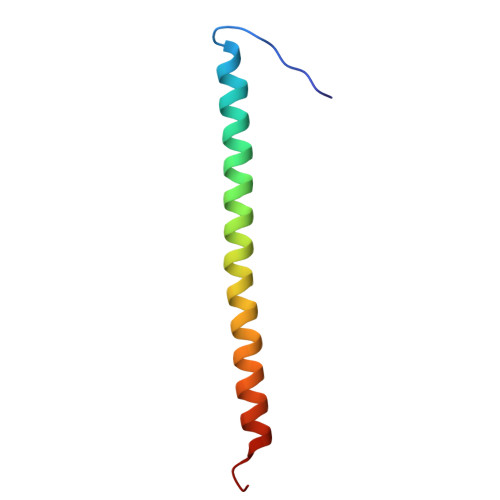
Assembly of subunit d (Vma6p) and G (Vma10p) and the NMR solution structure of subunit G (G(1-59)) of the Saccharomyces cerevisiae V(1)V(O) ATPase.
Rishikesan, S., Gayen, S., Thaker, Y.R., Vivekanandan, S., Manimekalai, M.S., Yau, Y.H., Shochat, S.G., Gruber, G.(2009) Biochim Biophys Acta 1787: 242-251
- PubMed: 19344662
- DOI: https://doi.org/10.1016/j.bbabio.2009.01.010
- Primary Citation of Related Structures:
2K88 - PubMed Abstract:
Understanding the structural traits of subunit G is essential, as it is needed for V(1)V(O) assembly and function. Here solution NMR of the recombinant N- (G(1-59)) and C-terminal segment (G(61-114)) of subunit G, has been performed in the absence and presence of subunit d of the yeast V-ATPase. The data show that G does bind to subunit d via its N-terminal part, G(1-59) only. The residues of G(1-59) involved in d binding are Gly7 to Lys34. The structure of G(1-59) has been solved, revealing an alpha-helix between residues 10 and 56, whereby the first nine- and the last three residues of G(1-59) are flexible. The surface charge distribution of G(1-59) reveals an amphiphilic character at the N-terminus due to positive and negative charge distribution at one side and a hydrophobic surface on the opposite side of the structure. The C-terminus exhibits a strip of negative residues. The data imply that G(1-59)-d assembly is accomplished by hydrophobic interactions and salt-bridges of the polar residues. Based on the recently determined NMR structure of segment E(18-38) of subunit E of yeast V-ATPase and the presently solved structure of G(1-59), both proteins have been docked and binding epitopes have been analyzed.
Organizational Affiliation:
School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, Singapore 637551, Republic of Singapore.