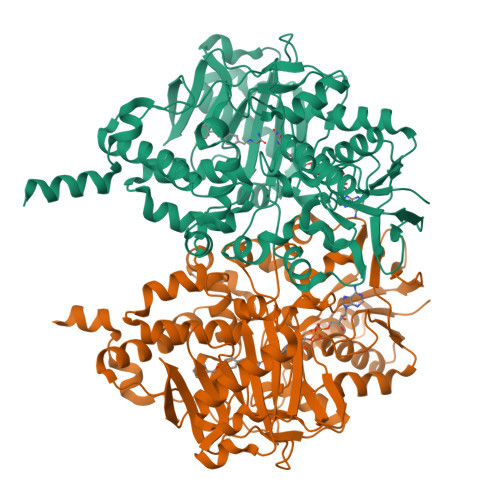
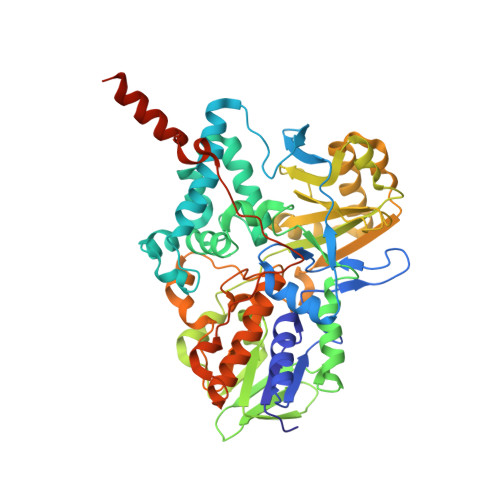
Structures of human monoamine oxidase B complexes with selective noncovalent inhibitors: safinamide and coumarin analogs.
Binda, C., Wang, J., Pisani, L., Caccia, C., Carotti, A., Salvati, P., Edmondson, D.E., Mattevi, A.(2007) J Med Chem 50: 5848-5852
- PubMed: 17915852
- DOI: https://doi.org/10.1021/jm070677y
- Primary Citation of Related Structures:
2V5Z, 2V60, 2V61 - PubMed Abstract:
Structures of human monoamine oxidase B (MAO B) in complex with safinamide and two coumarin derivatives, all sharing a common benzyloxy substituent, were determined by X-ray crystallography. These compounds competitively inhibit MAO B with Ki values in the 0.1-0.5 microM range that are 30-700-fold lower than those observed with MAO A. The inhibitors bind noncovalently to MAO B, occupying both the entrance and the substrate cavities and showing a similarly oriented benzyloxy substituent.
Organizational Affiliation:
Department of Genetics and Microbiology, University of Pavia, via Ferrata 1, Pavia, 27100 Italy.