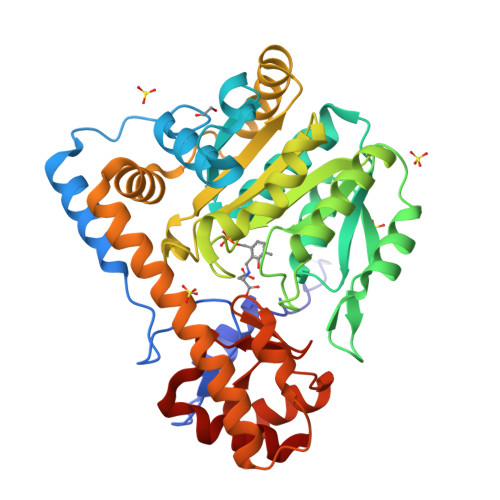
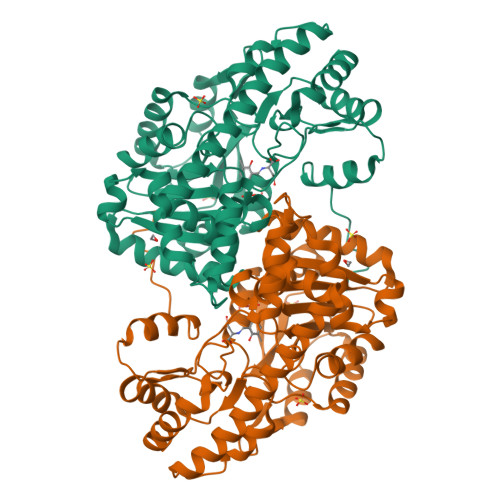
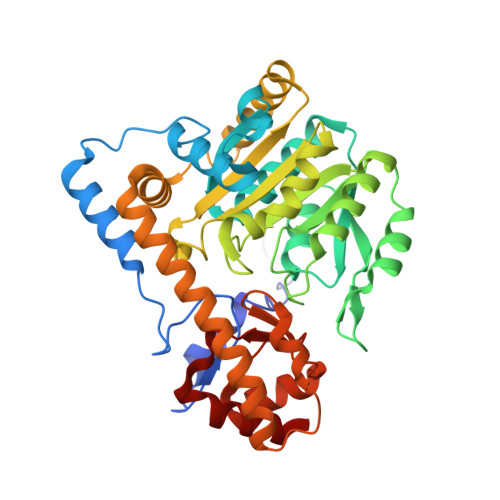
Ground-state electronic destabilization via hyperconjugation in aspartate aminotransferase.
Griswold, W.R., Castro, J.N., Fisher, A.J., Toney, M.D.(2012) J Am Chem Soc 134: 8436-8438
- PubMed: 22551424
- DOI: https://doi.org/10.1021/ja302809e
- Primary Citation of Related Structures:
4DBC - PubMed Abstract:
Binding isotope effects for l-aspartate reacting with the inactive K258A mutant of PLP-dependent aspartate aminotransferase to give a stable external aldimine intermediate are reported. They provide direct evidence for electronic ground-state destabilization via hyperconjugation. The smaller equilibrium isotope effect with deazaPLP-reconstituted K258A indicates that the pyridine nitrogen plays an important role in labilizing the Cα-H bond.
Organizational Affiliation:
Department of Chemistry, University of California - Davis, 95616, United States.