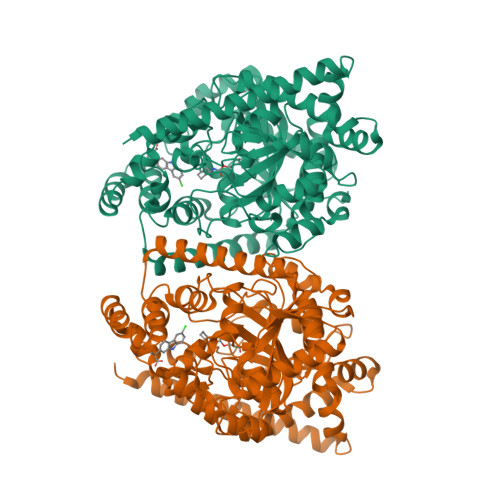
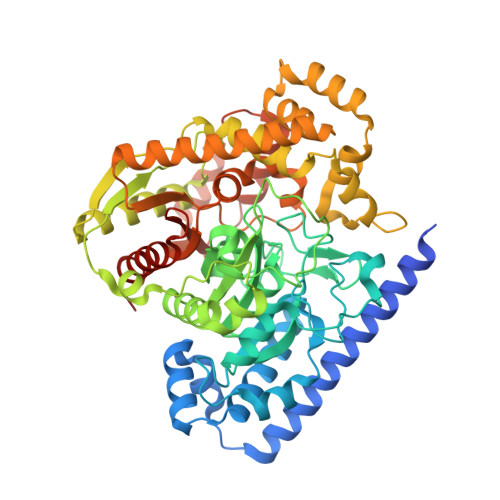
A Binding Site for Nonsteroidal Anti-inflammatory Drugs in Fatty Acid Amide Hydrolase.
Bertolacci, L., Romeo, E., Veronesi, M., Magotti, P., Albani, C., Dionisi, M., Lambruschini, C., Scarpelli, R., Cavalli, A., De Vivo, M., Piomelli, D., Garau, G.(2013) J Am Chem Soc 135: 22-25
- PubMed: 23240907
- DOI: https://doi.org/10.1021/ja308733u
- Primary Citation of Related Structures:
4DO3 - PubMed Abstract:
In addition to inhibiting the cyclooxygenase (COX)-mediated biosynthesis of prostanoids, various widely used nonsteroidal anti-inflammatory drugs (NSAIDs) enhance endocannabinoid signaling by blocking the anandamide-degrading membrane enzyme fatty acid amide hydrolase (FAAH). The X-ray structure of FAAH in complex with the NSAID carprofen, along with site-directed mutagenesis, enzyme activity assays, and NMR analysis, has revealed the molecular details of this interaction, providing information that may guide the design of dual FAAH-COX inhibitors with superior analgesic efficacy.
Organizational Affiliation:
Drug Discovery and Development, Istituto Italiano di Tecnologia, Via Morego 30, 16163 Genoa, Italy.