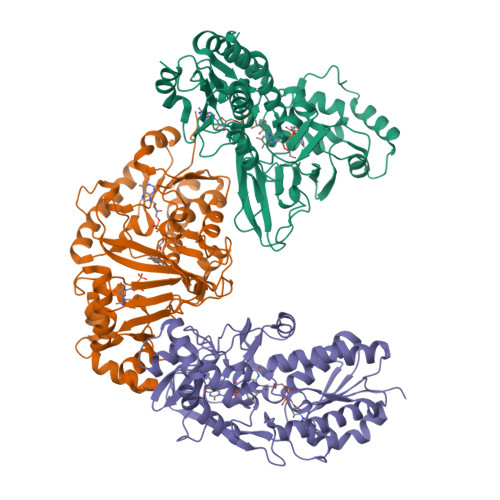
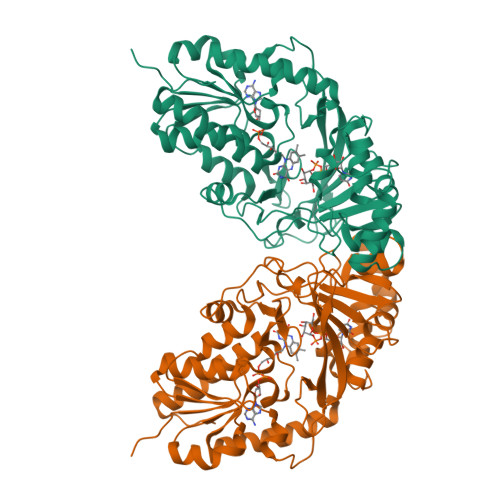
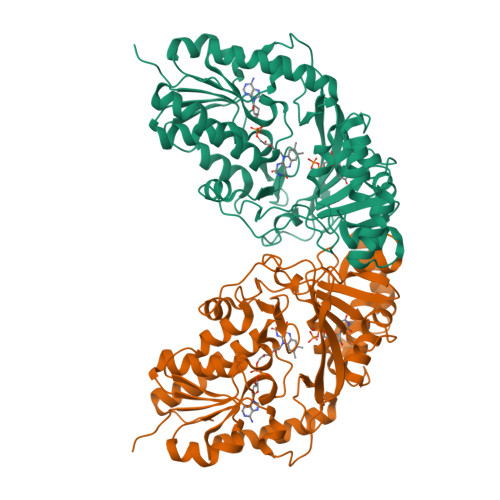
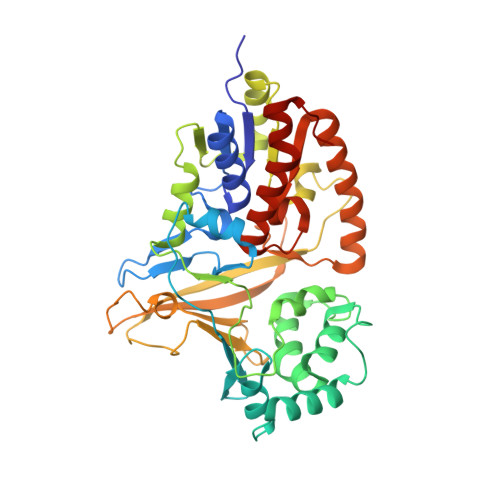
Structural Basis of Ligand Binding to UDP-Galactopyranose Mutase from Mycobacterium tuberculosis Using Substrate and Tetrafluorinated Substrate Analogues.
van Straaten, K.E., Kuttiyatveetil, J.R., Sevrain, C.M., Villaume, S.A., Jimenez-Barbero, J., Linclau, B., Vincent, S.P., Sanders, D.A.(2015) J Am Chem Soc 137: 1230-1244
- PubMed: 25562380
- DOI: https://doi.org/10.1021/ja511204p
- Primary Citation of Related Structures:
4RPG, 4RPH, 4RPJ, 4RPK, 4RPL - PubMed Abstract:
UDP-Galactopyranose mutase (UGM) is a flavin-containing enzyme that catalyzes the reversible conversion of UDP-galactopyranose (UDP-Galp) to UDP-galactofuranose (UDP-Galf) and plays a key role in the biosynthesis of the mycobacterial cell wall galactofuran. A soluble, active form of UGM from Mycobacterium tuberculosis (MtUGM) was obtained from a dual His6-MBP-tagged MtUGM construct. We present the first complex structures of MtUGM with bound substrate UDP-Galp (both oxidized flavin and reduced flavin). In addition, we have determined the complex structures of MtUGM with inhibitors (UDP and the dideoxy-tetrafluorinated analogues of both UDP-Galp (UDP-F4-Galp) and UDP-Galf (UDP-F4-Galf)), which represent the first complex structures of UGM with an analogue in the furanose form, as well as the first structures of dideoxy-tetrafluorinated sugar analogues bound to a protein. These structures provide detailed insight into ligand recognition by MtUGM and show an overall binding mode similar to those reported for other prokaryotic UGMs. The binding of the ligand induces conformational changes in the enzyme, allowing ligand binding and active-site closure. In addition, the complex structure of MtUGM with UDP-F4-Galf reveals the first detailed insight into how the furanose moiety binds to UGM. In particular, this study confirmed that the furanoside adopts a high-energy conformation ((4)E) within the catalytic pocket. Moreover, these investigations provide structural insights into the enhanced binding of the dideoxy-tetrafluorinated sugars compared to unmodified analogues. These results will help in the design of carbohydrate mimetics and drug development, and show the enormous possibilities for the use of polyfluorination in the design of carbohydrate mimetics.
Organizational Affiliation:
Department of Chemistry, University of Saskatchewan , 110 Science Place, Saskatoon S7N 5C9, Canada.