Target-Based Identification of Whole-Cell Active Inhibitors of Biotin Biosynthesis in Mycobacterium tuberculosis.
Park, S.W., Casalena, D.E., Wilson, D.J., Dai, R., Nag, P.P., Liu, F., Boyce, J.P., Bittker, J.A., Schreiber, S.L., Finzel, B.C., Schnappinger, D., Aldrich, C.C.(2015) Chem Biol 22: 76-86
- PubMed: 25556942
- DOI: https://doi.org/10.1016/j.chembiol.2014.11.012
- Primary Citation of Related Structures:
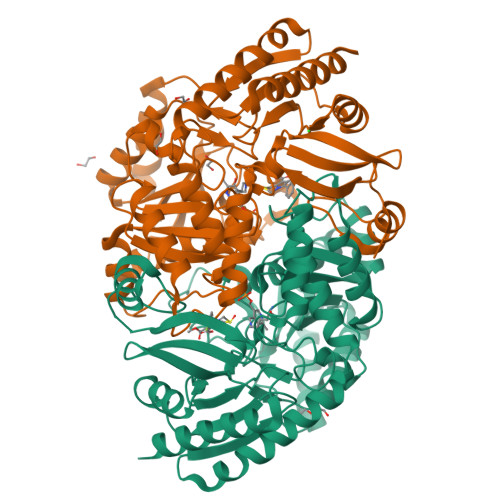
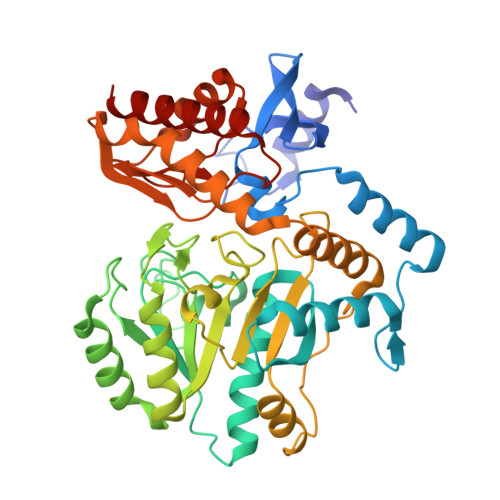
4W1V, 4W1W, 4W1X - PubMed Abstract:
Biotin biosynthesis is essential for survival and persistence of Mycobacterium tuberculosis (Mtb) in vivo. The aminotransferase BioA, which catalyzes the antepenultimate step in the biotin pathway, has been established as a promising target due to its vulnerability to chemical inhibition. We performed high-throughput screening (HTS) employing a fluorescence displacement assay and identified a diverse set of potent inhibitors including many diversity-oriented synthesis (DOS) scaffolds. To efficiently select only hits targeting biotin biosynthesis, we then deployed a whole-cell counterscreen in biotin-free and biotin-containing medium against wild-type Mtb and in parallel with isogenic bioA Mtb strains that possess differential levels of BioA expression. This counterscreen proved crucial to filter out compounds whose whole-cell activity was off target as well as identify hits with weak, but measurable whole-cell activity in BioA-depleted strains. Several of the most promising hits were cocrystallized with BioA to provide a framework for future structure-based drug design efforts.
Organizational Affiliation:
Department of Microbiology and Immunology, Weill Cornell Medical College, New York, NY 10065, USA.