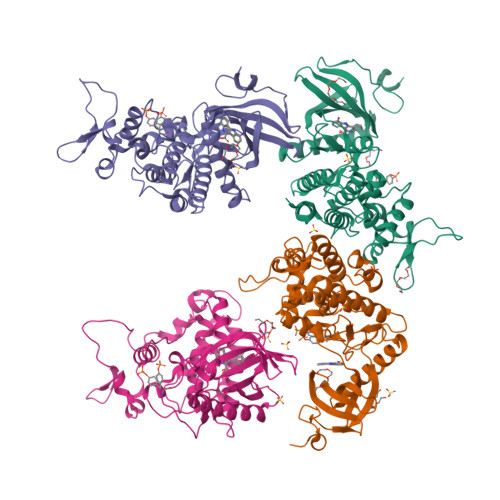
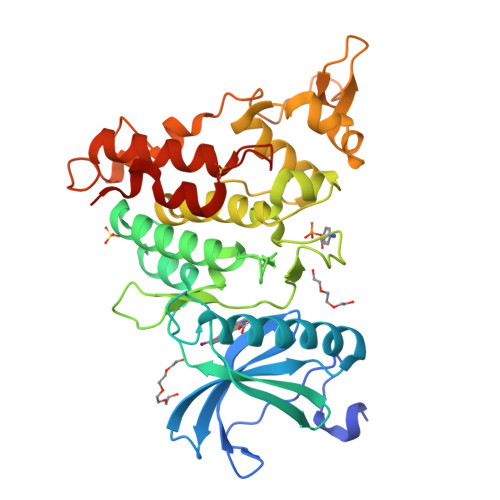
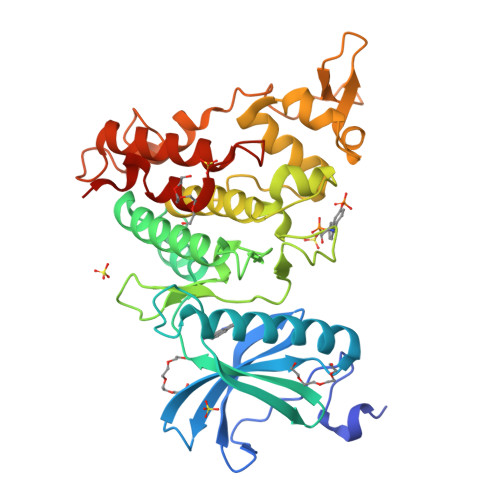
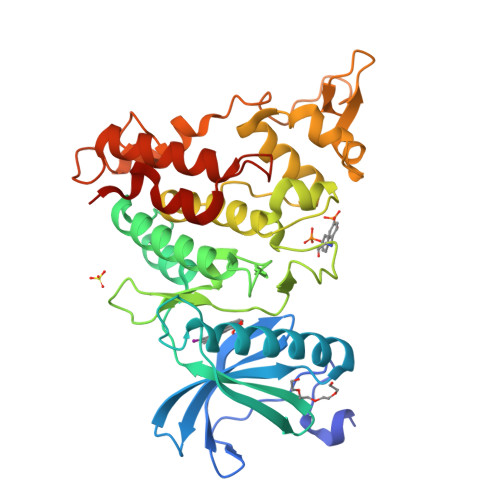
10-Iodo-11H-indolo[3,2-c]quinoline-6-carboxylic Acids Are Selective Inhibitors of DYRK1A.
Falke, H., Chaikuad, A., Becker, A., Loaec, N., Lozach, O., Abu Jhaisha, S., Becker, W., Jones, P.G., Preu, L., Baumann, K., Knapp, S., Meijer, L., Kunick, C.(2015) J Med Chem 58: 3131-3143
- PubMed: 25730262
- DOI: https://doi.org/10.1021/jm501994d
- Primary Citation of Related Structures:
4YLJ, 4YLK, 4YLL - PubMed Abstract:
The protein kinase DYRK1A has been suggested to act as one of the intracellular regulators contributing to neurological alterations found in individuals with Down syndrome. For an assessment of the role of DYRK1A, selective synthetic inhibitors are valuable pharmacological tools. However, the DYRK1A inhibitors described in the literature so far either are not sufficiently selective or have not been tested against closely related kinases from the DYRK and the CLK protein kinase families. The aim of this study was the identification of DYRK1A inhibitors exhibiting selectivity versus the structurally and functionally closely related DYRK and CLK isoforms. Structure modification of the screening hit 11H-indolo[3,2-c]quinoline-6-carboxylic acid revealed structure-activity relationships for kinase inhibition and enabled the design of 10-iodo-substituted derivatives as very potent DYRK1A inhibitors with considerable selectivity against CLKs. X-ray structure determination of three 11H-indolo[3,2-c]quinoline-6-carboxylic acids cocrystallized with DYRK1A confirmed the predicted binding mode within the ATP binding site.
Organizational Affiliation:
†Institut für Medizinische und Pharmazeutische Chemie, Technische Universität Braunschweig, Beethovenstraße 55, 38106 Braunschweig, Germany.