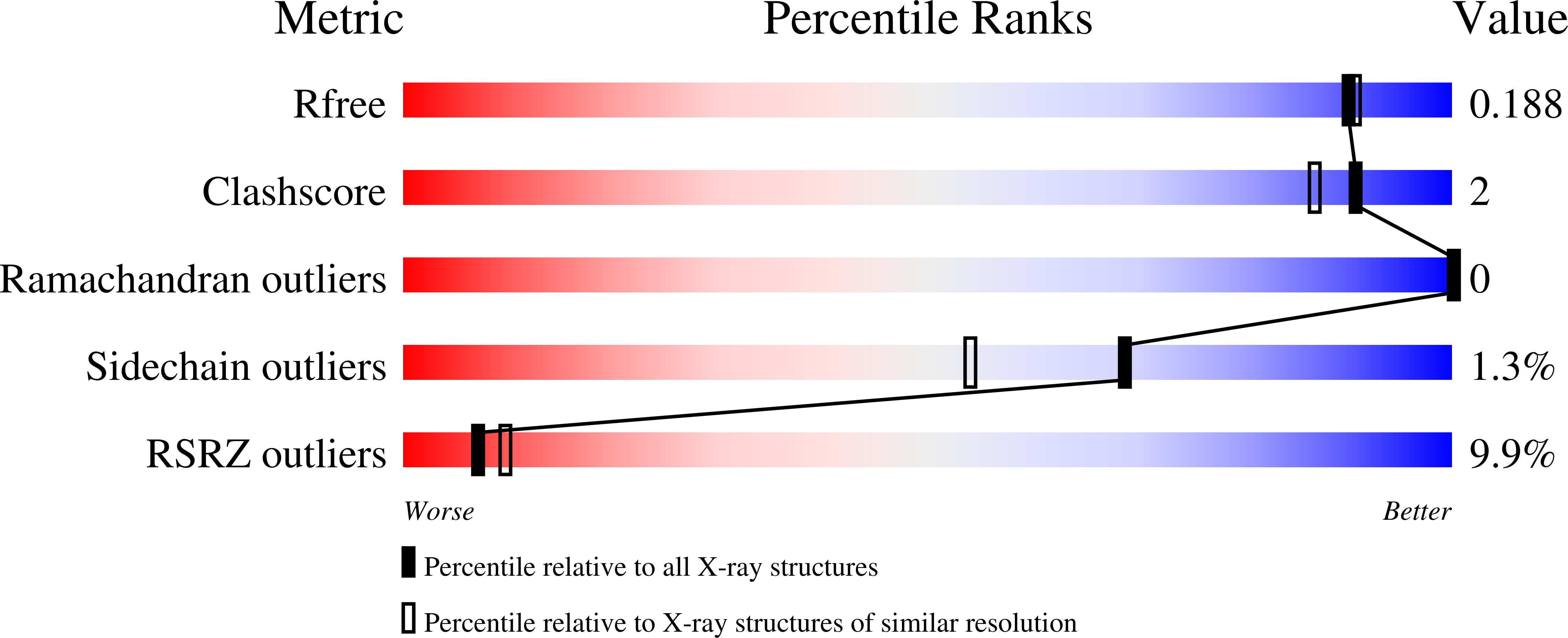
Targeting Bacterial Nitric Oxide Synthase with Aminoquinoline-Based Inhibitors.
Holden, J.K., Lewis, M.C., Cinelli, M.A., Abdullatif, Z., Pensa, A.V., Silverman, R.B., Poulos, T.L.(2016) Biochemistry 55: 5587-5594
- PubMed: 27607918
- DOI: https://doi.org/10.1021/acs.biochem.6b00786
- Primary Citation of Related Structures:
5G65, 5G66, 5G67, 5G68, 5G69, 5G6A, 5G6B, 5G6C, 5G6D, 5G6E, 5G6F, 5G6G, 5G6H, 5G6I, 5G6J, 5G6K, 5G6L, 5G6M, 5G6N, 5G6O, 5G6P, 5G6Q - PubMed Abstract:
Nitric oxide is produced in Gram-positive pathogens Bacillus anthracis and Staphylococcus aureus by the bacterial isoform of nitric oxide synthase (NOS). Inhibition of bacterial nitric oxide synthase (bNOS) has been identified as a promising antibacterial strategy for targeting methicillin-resistant S. aureus [Holden, J. K., et al. (2015) Chem. Biol. 22, 785-779]. One class of NOS inhibitors that demonstrates antimicrobial efficacy utilizes an aminoquinoline scaffold. Here we report on a variety of aminoquinolines that target the bacterial NOS active site, in part, by binding to a hydrophobic patch that is unique to bNOS. Through mutagenesis and crystallographic studies, our findings demonstrate that aminoquinolines are an excellent scaffold for further aiding in the development of bNOS specific inhibitors.
Organizational Affiliation:
Department of Molecular Biology and Biochemistry, ‡Department of Pharmaceutical Sciences, and §Department of Chemistry, University of California , Irvine, California 92697-3900, United States.