Discovery of N-substituted 7-azaindoles as PIM1 kinase inhibitors - Part I.
Barberis, C., Moorcroft, N., Arendt, C., Levit, M., Moreno-Mazza, S., Batchelor, J., Mechin, I., Majid, T.(2017) Bioorg Med Chem Lett 27: 4730-4734
- PubMed: 28947155
- DOI: https://doi.org/10.1016/j.bmcl.2017.08.069
- Primary Citation of Related Structures:
5KCX, 5TEL, 5TEX, 5TOE, 5TUR - PubMed Abstract:
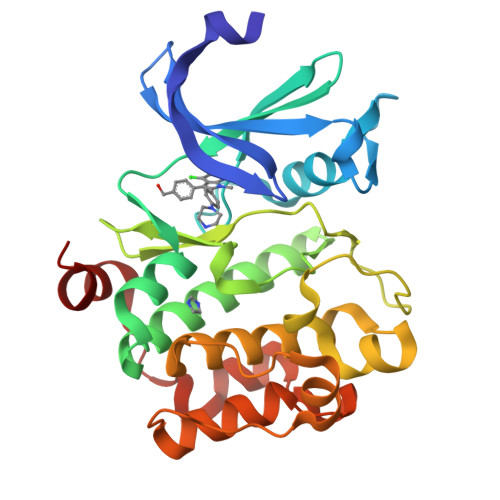
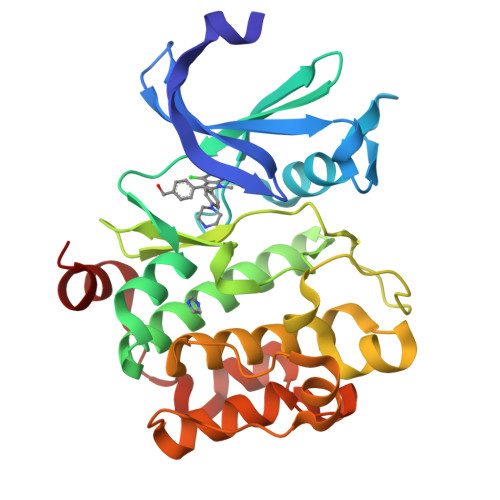
Novel N-substituted azaindoles have been discovered as PIM1 inhibitors. X-ray structures have played a significant role in orienting the chemistry effort in the initial phase of hit confirmation. Disclosure of an unconventional binding mode for 1 and 2, as demonstrated by X-ray crystallography, is presented and was an important factor in selecting and advancing a lead series.
Organizational Affiliation:
IDD Medicinal Chemistry, Sanofi Genzyme, 153 Second Avenue, Waltham MA 02451, USA. Electronic address: claude.barberis2@sanofi.com.