Synthesis, Antiviral Potency, in Vitro ADMET, and X-ray Structure of Potent CD4 Mimics as Entry Inhibitors That Target the Phe43 Cavity of HIV-1 gp120.
Curreli, F., Kwon, Y.D., Belov, D.S., Ramesh, R.R., Kurkin, A.V., Altieri, A., Kwong, P.D., Debnath, A.K.(2017) J Med Chem 60: 3124-3153
- PubMed: 28266845
- DOI: https://doi.org/10.1021/acs.jmedchem.7b00179
- Primary Citation of Related Structures:
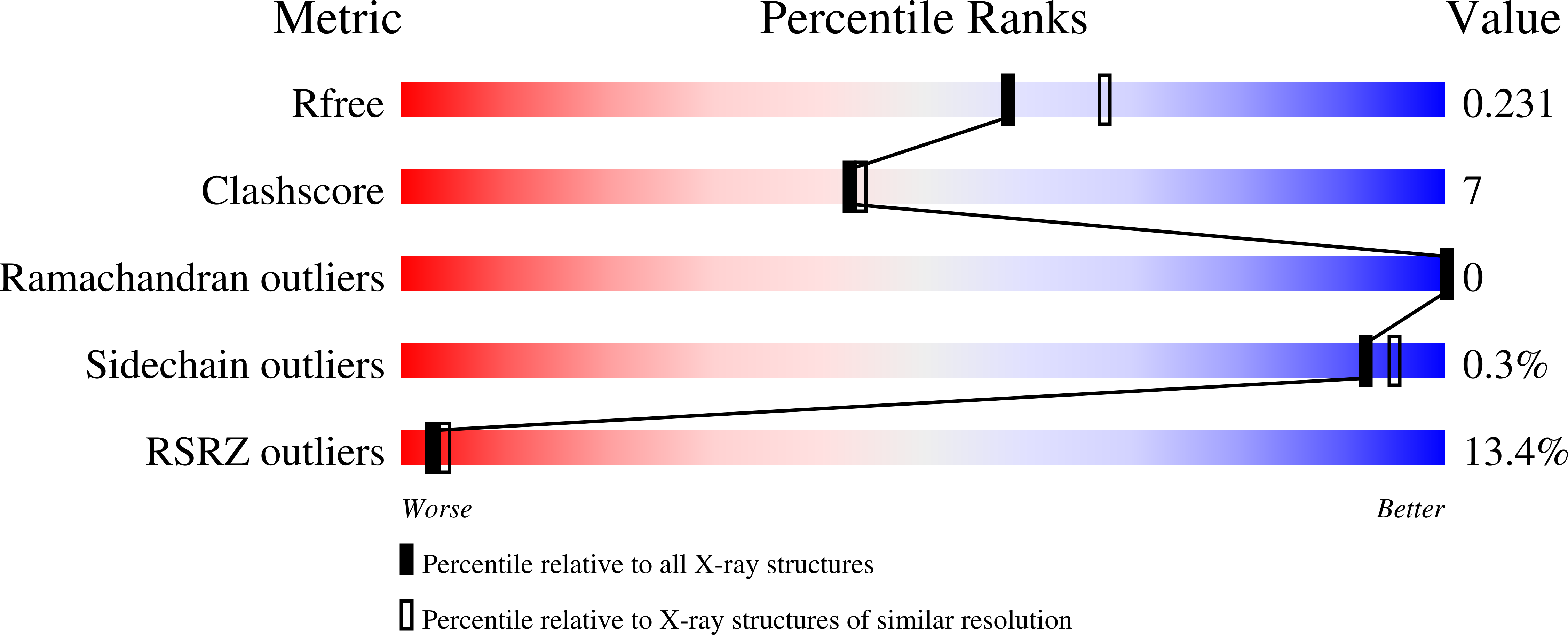
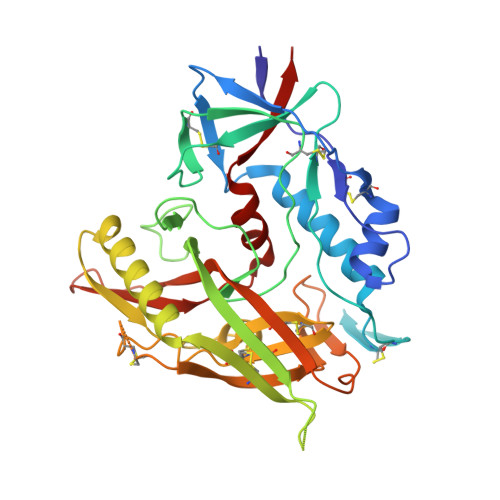
5U6E - PubMed Abstract:
In our attempt to optimize the lead HIV-1 entry antagonist, NBD-11021, we present in this study the rational design and synthesis of 60 new analogues and determination of their antiviral activity in a single-cycle and a multicycle infection assay to derive a comprehensive structure-activity relationship (SAR). Two of these compounds, NBD-14088 and NBD-14107, showed significant improvement in antiviral activity compared to the lead entry antagonist in a single-cycle assay against a large panel of Env-pseudotyped viruses. The X-ray structure of a similar compound, NBD-14010, confirmed the binding mode of the newly designed compounds. The in vitro ADMET profiles of these compounds are comparable to that of the most potent attachment inhibitor BMS-626529, a prodrug of which is currently undergoing phase III clinical trials. The systematic study presented here is expected to pave the way for improving the potency, toxicity, and ADMET profile of this series of compounds with the potential to be moved to the early preclinical development.
Organizational Affiliation:
Laboratory of Molecular Modeling and Drug Design, Lindsey F. Kimball Research Institute, New York Blood Center , 310 E 67th Street, New York, New York 10065, United States.