Atg7 Activates an Autophagy-Essential Ubiquitin-like Protein Atg8 through Multi-Step Recognition.
Yamaguchi, M., Satoo, K., Suzuki, H., Fujioka, Y., Ohsumi, Y., Inagaki, F., Noda, N.N.(2018) J Mol Biology 430: 249-257
- PubMed: 29237558
- DOI: https://doi.org/10.1016/j.jmb.2017.12.002
- Primary Citation of Related Structures:
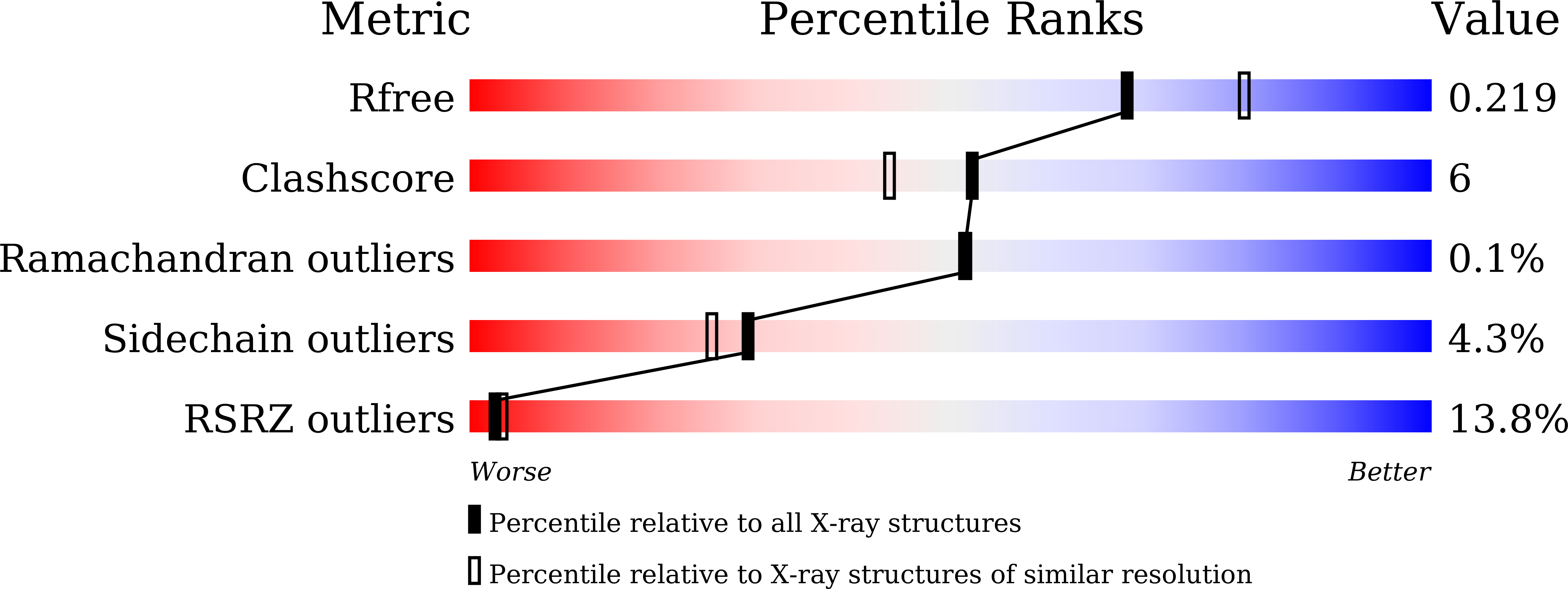
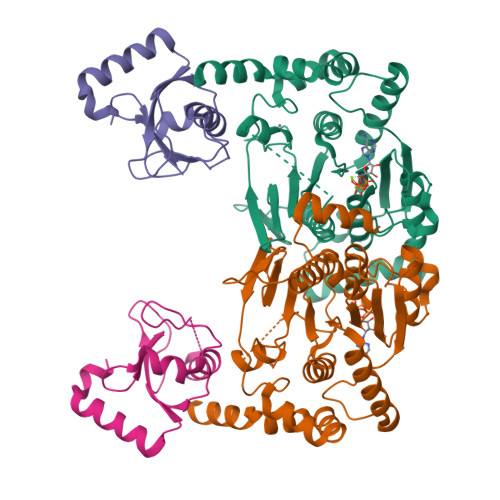
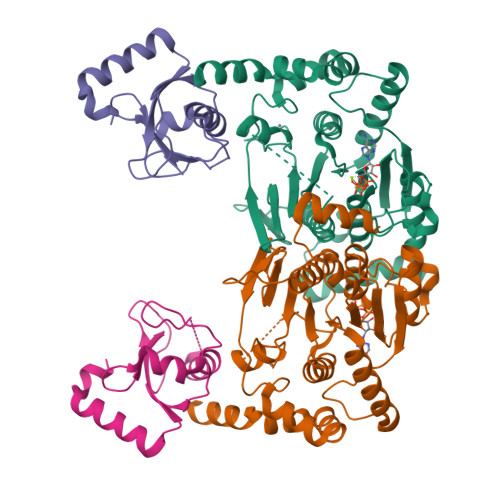
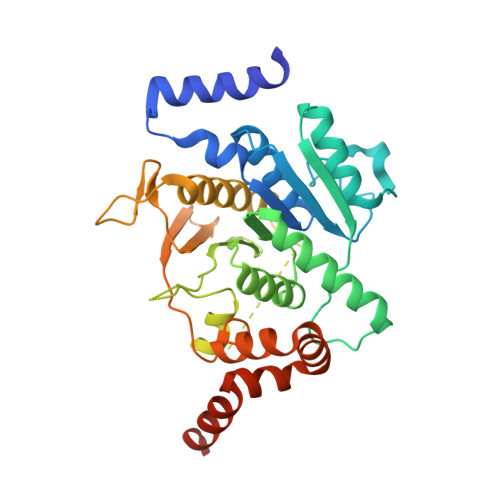
5YEC - PubMed Abstract:
Atg8 is a unique ubiquitin-like protein that is covalently conjugated with a phosphatidylethanolamine through reactions similar to ubiquitination and plays essential roles in autophagy. Atg7 is the E1 enzyme for Atg8, and it activates the C-terminal Gly116 of Atg8 using ATP. Here, we report the crystal structure of Atg8 bound to the C-terminal domain of Atg7 in an unprecedented mode. Atg8 neither contacts with the central β-sheet nor binds to the catalytic site of Atg7, both of which were observed in previously reported Atg7-Atg8 structures. Instead, Atg8 binds to the C-terminal α-helix and crossover loop, thereby changing the autoinhibited conformation of the crossover loop observed in the free Atg7 structure into a short helix and a disordered loop. Mutational analyses suggested that this interaction mode is important for the activation reaction. We propose that Atg7 recognizes Atg8 through multiple steps, which would be necessary to induce a conformational change in Atg7 that is optimal for the activation reaction.
Organizational Affiliation:
Department of Structural Biology, Faculty of Advanced Life Science, Hokkaido University, Sapporo 060-0810, Japan.