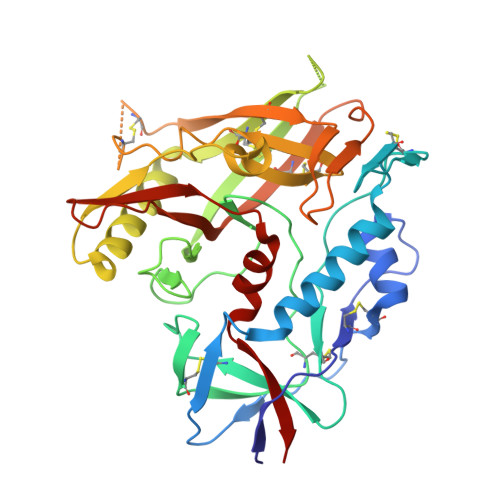
A New Family of Small-Molecule CD4-Mimetic Compounds Contacts Highly Conserved Aspartic Acid 368 of HIV-1 gp120 and Mediates Antibody-Dependent Cellular Cytotoxicity.
Ding, S., Grenier, M.C., Tolbert, W.D., Vezina, D., Sherburn, R., Richard, J., Prevost, J., Chapleau, J.P., Gendron-Lepage, G., Medjahed, H., Abrams, C., Sodroski, J., Pazgier, M., Smith 3rd, A.B., Finzi, A.(2019) J Virol 93
- PubMed: 31554684
- DOI: https://doi.org/10.1128/JVI.01325-19
- Primary Citation of Related Structures:
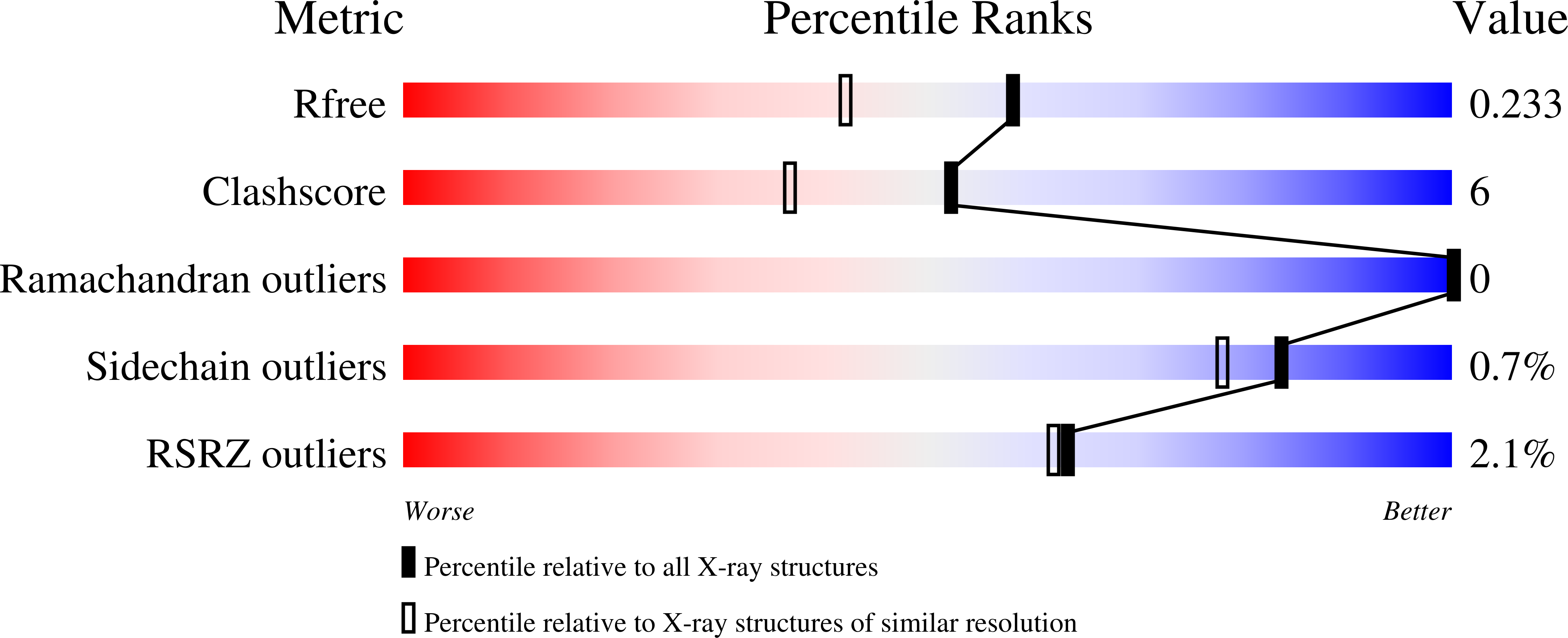
6ONE, 6ONF, 6ONH, 6ONV, 6P9N - PubMed Abstract:
The HIV-1 envelope glycoprotein (Env) trimer mediates virus entry into cells. The "closed" conformation of Env is resistant to nonneutralizing antibodies (nnAbs). These antibodies mostly recognize occluded epitopes that can be exposed upon binding of CD4 or small-molecule CD4 mimetics (CD4mc). Here, we describe a new family of small molecules that expose Env to nnAbs and sensitize infected cells to antibody-dependent cellular cytotoxicity (ADCC). These compounds have a limited capacity to inhibit virus infection directly but are able to sensitize viral particles to neutralization by otherwise nonneutralizing antibodies. Structural analysis shows that some analogs of this family of CD4mc engage the gp120 Phe43 cavity by contacting the highly conserved D368 residue, making them attractive scaffolds for drug development. IMPORTANCE HIV-1 has evolved multiple strategies to avoid humoral responses. One efficient mechanism is to keep its envelope glycoprotein (Env) in its "closed" conformation. Here, we report on a new family of small molecules that are able to "open up" Env, thus exposing vulnerable epitopes. This new family of molecules binds in the Phe43 cavity and contacts the highly conserved D368 residue. The structural and biological attributes of molecules of this family make them good candidates for drug development.
Organizational Affiliation:
Centre de Recherche du CHUM, Montreal, Quebec, Canada.