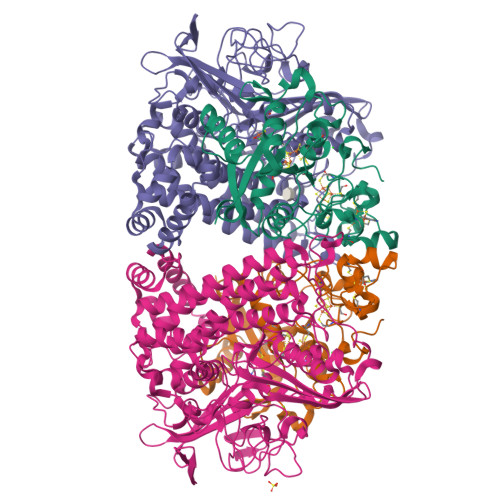
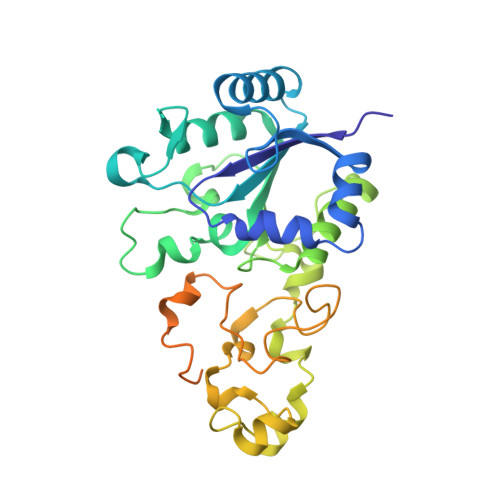
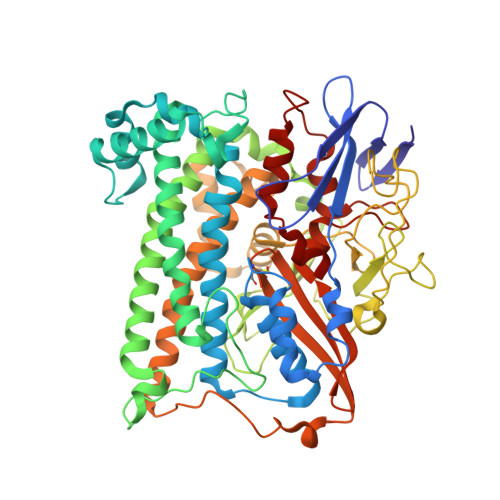
Aerobic Photocatalytic H2Production by a [NiFe] Hydrogenase Engineered to Place a Silver Nanocluster in the Electron Relay.
Zhang, L., Morello, G., Carr, S.B., Armstrong, F.A.(2020) J Am Chem Soc 142: 12699-12707
- PubMed: 32579353
- DOI: https://doi.org/10.1021/jacs.0c04302
- Primary Citation of Related Structures:
6RP2 - PubMed Abstract:
Hydrogenase-1 (Hyd-1) from E. coli poses a conundrum regarding the properties of electrocatalytic reversibility and associated bidirectionality now established for many redox enzymes. Its excellent H 2 -oxidizing activity begins only once a substantial overpotential is applied, and it cannot produce H 2 . A major reason for its unidirectional behavior is that the reduction potentials of its electron-relaying FeS clusters are too positive relative to the 2H + /H 2 couple at neutral pH; consequently, electrons held within the enzyme lack the energy to drive H 2 production. However, Hyd-1 is O 2 -tolerant and even functions in air. Changing a tyrosine (Y) or threonine (T), located on the protein surface within 10 Å of the distal [4Fe-4S] and medial [3Fe-4S] clusters, to cysteine (C), allows site-selective attachment of a silver nanocluster (AgNC), the reduced or photoexcited state of which is a powerful reductant. The AgNC provides a new additional redox site, capturing externally supplied electrons with sufficiently high energy to drive H 2 production. Assemblies of Y'227C (or T'225C) with AgNCs/PMAA (PMAA = polymethyl acrylate templating several AgNC) are also electroactive for H 2 production at a TiO 2 electrode. A colloidal system for visible-light photo-H 2 generation is made by building the hybrid enzyme into a heterostructure with TiO 2 and graphitic carbon nitride (g-C 3 N 4 ), the resulting scaffold promoting uptake of electrons excited at the AgNC. Each hydrogenase produces 40 molecules of H 2 per second and sustains 20% activity in air.
Organizational Affiliation:
Inorganic Chemistry Laboratory, Department of Chemistry, University of Oxford, South Parks Road, Oxford OX1 3QR, Oxfordshire United Kingdom.