Substitution of the axial Type 1 Cu Ligand Afford Binding of a Water Molecule in Axial Position Affecting Kinetics, Spectral, and Structural Properties of the Small Laccase Ssl1.
Olbrich, A.C., Mielenbrink, S., Willers, V.P., Koschorreck, K., Birrell, J.A., Span, I., Urlacher, V.B.(2024) Chemistry : e202403005-e202403005
- PubMed: 39541228
- DOI: https://doi.org/10.1002/chem.202403005
- Primary Citation of Related Structures:
6Y4A, 6YO5, 6YZD, 6YZF, 6YZY - PubMed Abstract:
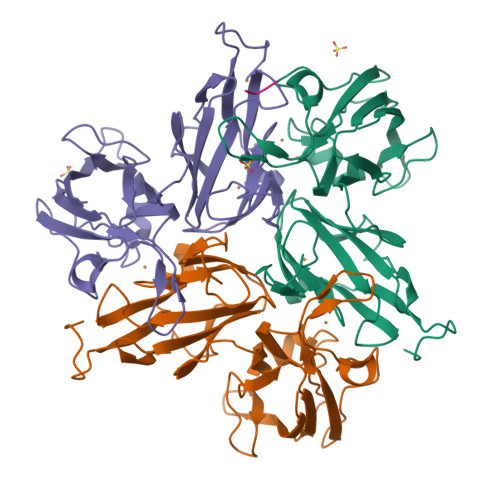
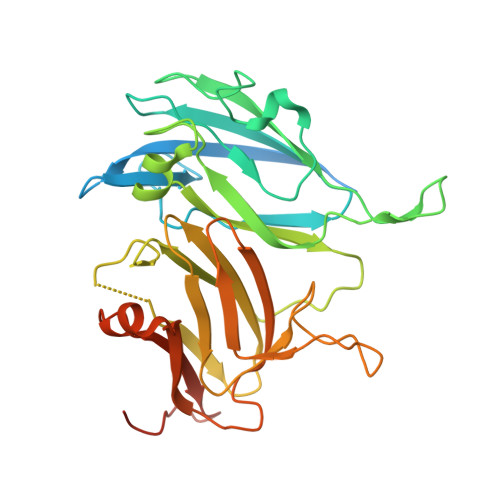
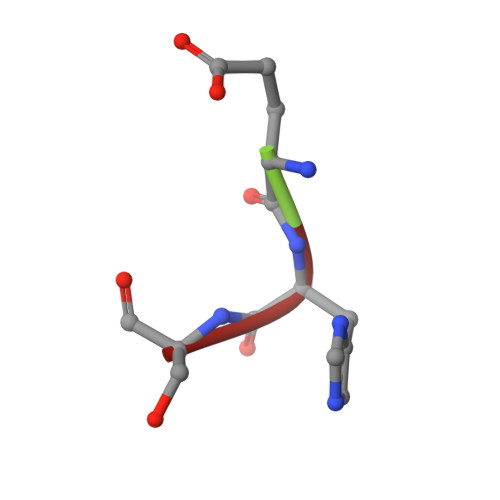
Multicopper oxidases use Cu ions as cofactors to oxidize various substrates. High reduction potential at Type 1 Cu is considered as crucial for effective catalysis. Previous studies have shown that replacing the axial methionine ligand of the Type 1 Cu with leucine or phenylalanine leads to an increased reduction potential, but not always to higher enzyme activity. Here we present a study on six variants of the small laccase Ssl1 from Streptomyces sviceus, where the axial methionine ligand was substituted, and the effect of the axial ligand on reduction potential, activity, spectral properties and structure was investigated. Absorption, electronic circular dichroism and EPR spectra revealed the presence of a stronger coordinating axial ligand like oxygen, which influences the electronic and catalytic properties more than the nature of the amino acid side chain. The crystal structures of the Ssl1 variants were solved, which show that none of the amino acid side chains coordinate to the Cu. Instead, a water molecule is found in the axial coordination site, which support the spectroscopic data. Our findings highlight the importance of combining structural and spectroscopic methods to investigate the effect of amino acid exchange on multicopper oxidases.
Organizational Affiliation:
Institute of Biochemistry, Heinrich Heine University Düsseldorf, Universitätsstr. 1, Düsseldorf, 40225, Germany.