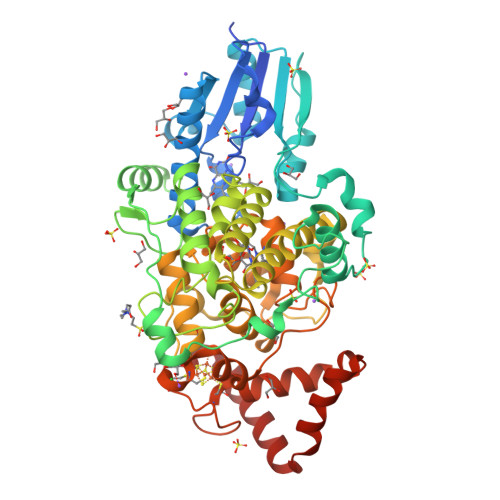
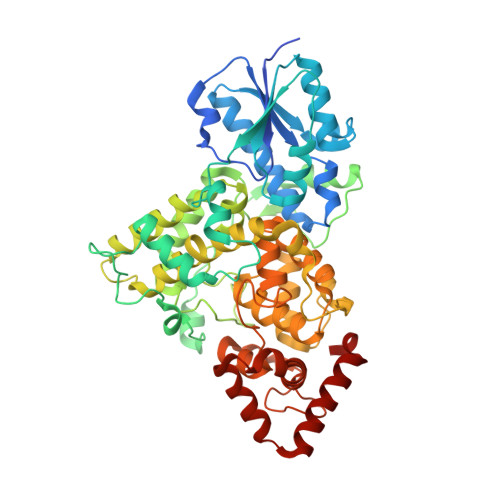
Structural and Functional Analysis of a Prokaryotic (6-4) Photolyase from the Aquatic Pathogen Vibrio Cholerae † .
Emmerich, H.J., Schneider, L., Essen, L.O.(2023) Photochem Photobiol 99: 1248-1257
- PubMed: 36692077
- DOI: https://doi.org/10.1111/php.13783
- Primary Citation of Related Structures:
8A1H - PubMed Abstract:
Photolyases are flavoproteins, which are able to repair UV-induced DNA lesions in a light-dependent manner. According to their substrate, they can be distinguished as CPD- and (6-4) photolyases. While CPD-photolyases repair the predominantly occurring cyclobutane pyrimidine dimer lesion, (6-4) photolyases catalyze the repair of the less prominent (6-4) photoproduct. The subgroup of prokaryotic (6-4) photolyases/FeS-BCP is one of the most ancient types of flavoproteins in the ubiquitously occurring photolyase & cryptochrome superfamily (PCSf). In contrast to canonical photolyases, prokaryotic (6-4) photolyases possess a few particular characteristics, including a lumazine derivative as antenna chromophore besides the catalytically essential flavin adenine dinucleotide as well as an elongated linker region between the N-terminal α/β-domain and the C-terminal all-α-helical domain. Furthermore, they can harbor an additional short subdomain, located at the C-terminus, with a binding site for a [4Fe-4S] cluster. So far, two crystal structures of prokaryotic (6-4) photolyases have been reported. Within this study, we present the high-resolution structure of the prokaryotic (6-4) photolyase from Vibrio cholerae and its spectroscopic characterization in terms of in vitro photoreduction and DNA-repair activity.
Organizational Affiliation:
Unit for Structural Biochemistry, Department of Chemistry, Philipps University Marburg, Marburg, Germany.