Stable Mammalian Serum Albumins Designed for Bacterial Expression.
Khersonsky, O., Goldsmith, M., Zaretsky, I., Hamer-Rogotner, S., Dym, O., Unger, T., Yona, M., Fridmann-Sirkis, Y., Fleishman, S.J.(2023) J Mol Biol 435: 168191-168191
- PubMed: 37385581
- DOI: https://doi.org/10.1016/j.jmb.2023.168191
- Primary Citation of Related Structures:
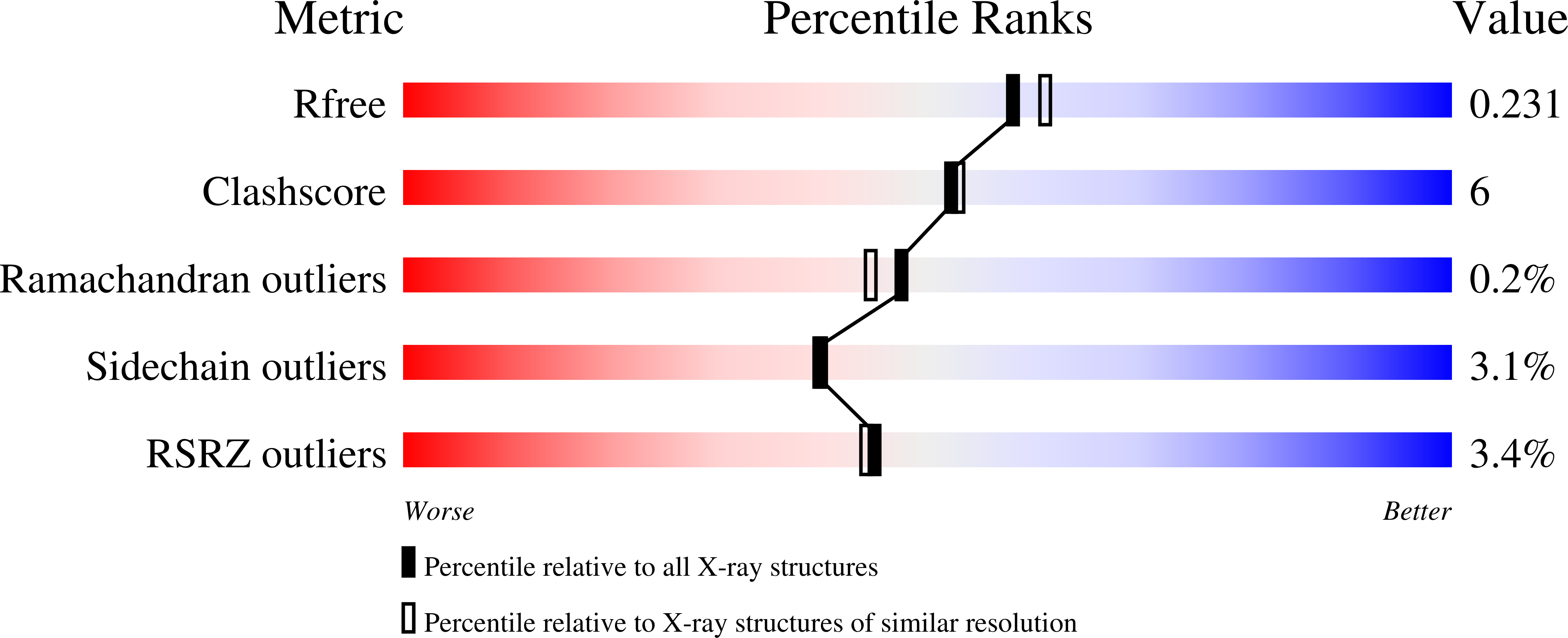
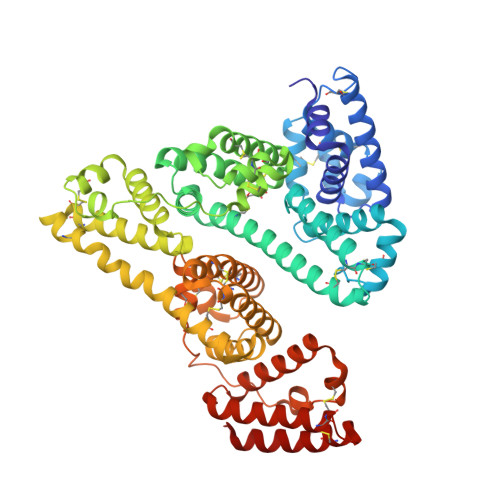
8A9Q - PubMed Abstract:
Albumin is the most abundant protein in the blood serum of mammals and has essential carrier and physiological roles. Albumins are also used in a wide variety of molecular and cellular experiments and in the cultivated meat industry. Despite their importance, however, albumins are challenging for heterologous expression in microbial hosts, likely due to 17 conserved intramolecular disulfide bonds. Therefore, albumins used in research and biotechnological applications either derive from animal serum, despite severe ethical and reproducibility concerns, or from recombinant expression in yeast or rice. We use the PROSS algorithm to stabilize human and bovine serum albumins, finding that all are highly expressed in E. coli. Design accuracy is verified by crystallographic analysis of a human albumin variant with 16 mutations. This albumin variant exhibits ligand binding properties similar to those of the wild type. Remarkably, a design with 73 mutations relative to human albumin exhibits over 40 °C improved stability and is stable beyond the boiling point of water. Our results suggest that proteins with many disulfide bridges have the potential to exhibit extreme stability when subjected to design. The designed albumins may be used to make economical, reproducible, and animal-free reagents for molecular and cell biology. They also open the way to high-throughput screening to study and enhance albumin carrier properties.
Organizational Affiliation:
Department of Biomolecular Sciences, Weizmann Institute of Science, Rehovot 7610001, Israel. Electronic address: [email protected].