Development of allosteric and selective CDK2 inhibitors for contraception with negative cooperativity to cyclin binding.
Faber, E.B., Sun, L., Tang, J., Roberts, E., Ganeshkumar, S., Wang, N., Rasmussen, D., Majumdar, A., Hirsch, L.E., John, K., Yang, A., Khalid, H., Hawkinson, J.E., Levinson, N.M., Chennathukuzhi, V., Harki, D.A., Schonbrunn, E., Georg, G.I.(2023) Nat Commun 14: 3213-3213
- PubMed: 37270540
- DOI: https://doi.org/10.1038/s41467-023-38732-x
- Primary Citation of Related Structures:
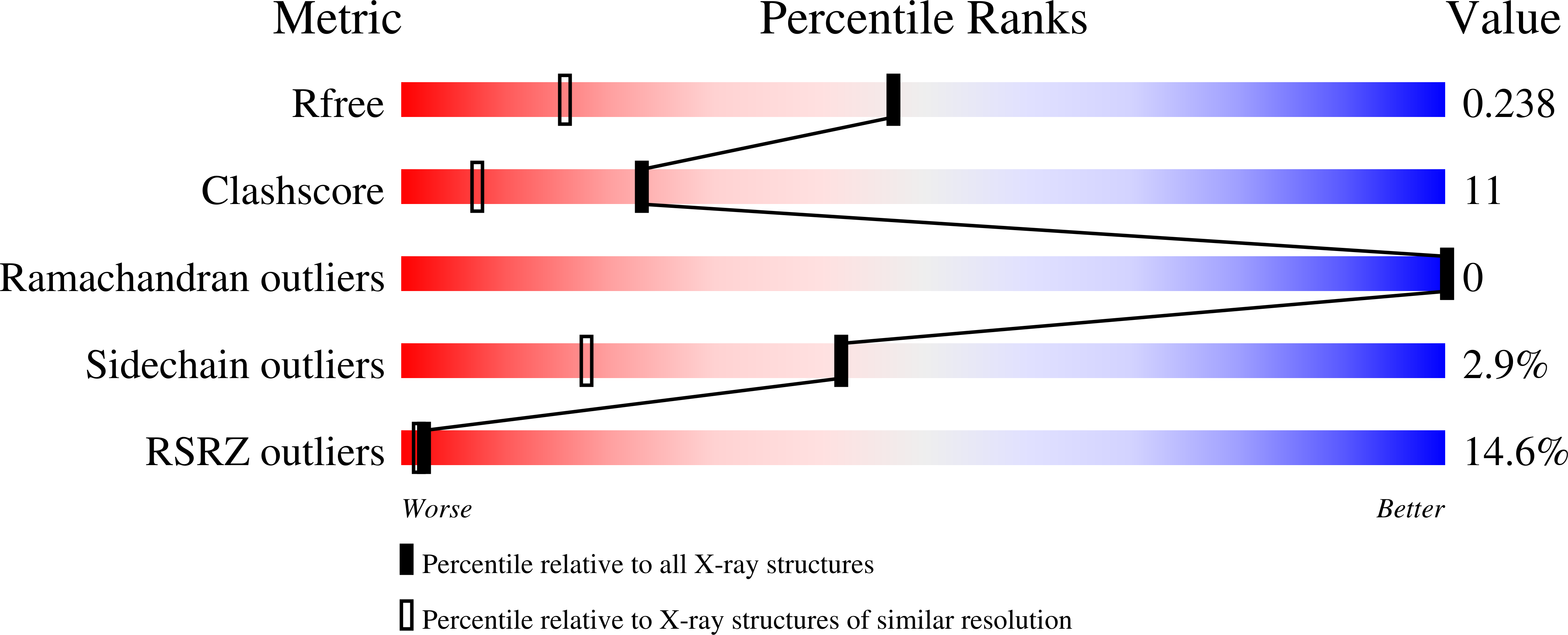
7RWF, 7S84, 8FOW, 8FP0, 8FP5 - PubMed Abstract:
Compared to most ATP-site kinase inhibitors, small molecules that target an allosteric pocket have the potential for improved selectivity due to the often observed lower structural similarity at these distal sites. Despite their promise, relatively few examples of structurally confirmed, high-affinity allosteric kinase inhibitors exist. Cyclin-dependent kinase 2 (CDK2) is a target for many therapeutic indications, including non-hormonal contraception. However, an inhibitor against this kinase with exquisite selectivity has not reached the market because of the structural similarity between CDKs. In this paper, we describe the development and mechanism of action of type III inhibitors that bind CDK2 with nanomolar affinity. Notably, these anthranilic acid inhibitors exhibit a strong negative cooperative relationship with cyclin binding, which remains an underexplored mechanism for CDK2 inhibition. Furthermore, the binding profile of these compounds in both biophysical and cellular assays demonstrate the promise of this series for further development into a therapeutic selective for CDK2 over highly similar kinases like CDK1. The potential of these inhibitors as contraceptive agents is seen by incubation with spermatocyte chromosome spreads from mouse testicular explants, where they recapitulate Cdk2 -/- and Spdya -/- phenotypes.
Organizational Affiliation:
Department of Medicinal Chemistry, University of Minnesota College of Pharmacy-Twin Cities, Minneapolis, MN, USA.