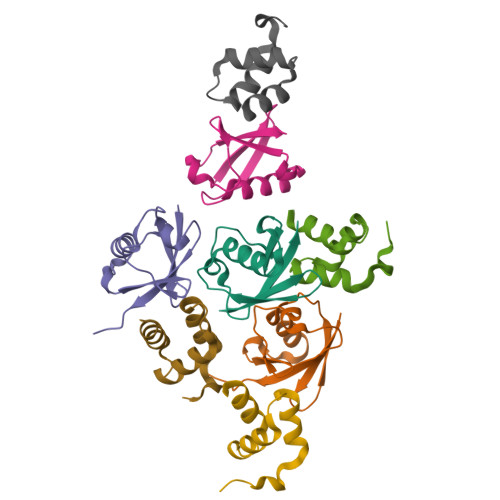
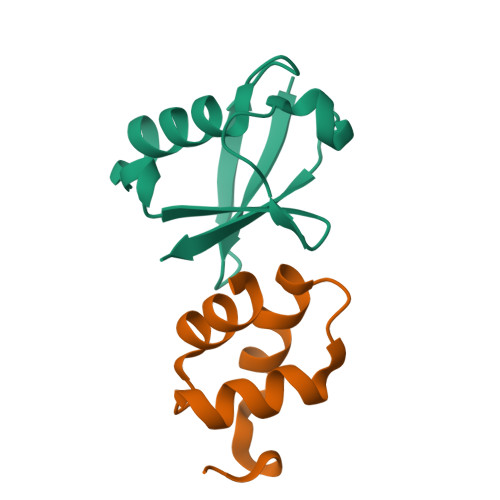
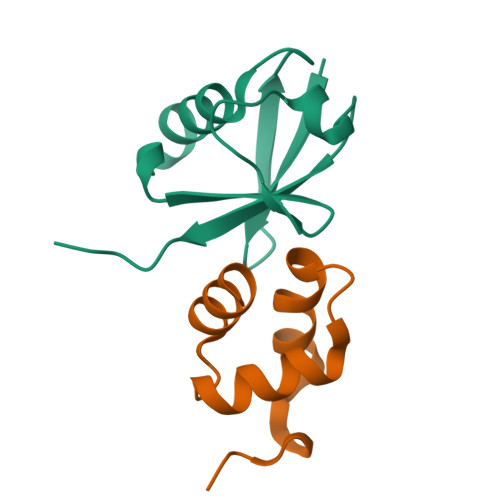
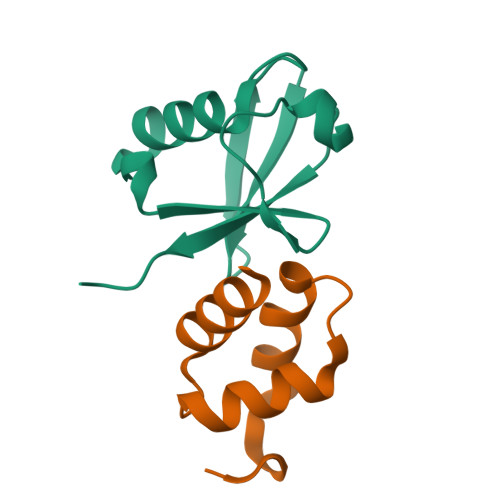
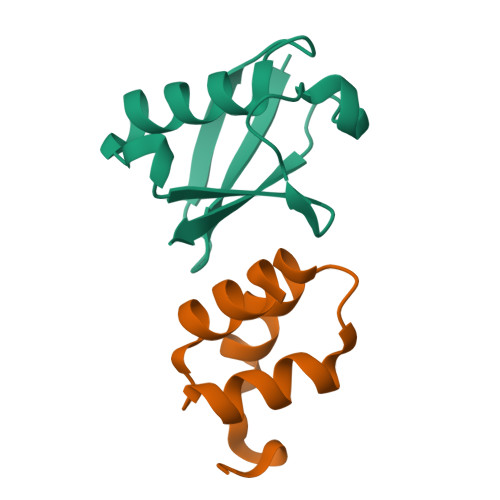
Structural basis of ubiquitin recognition by the ubiquitin-associated (UBA) domain of the ubiquitin ligase EDD.
Kozlov, G., Nguyen, L., Lin, T., De Crescenzo, G., Park, M., Gehring, K.(2007) J Biological Chem 282: 35787-35795
- PubMed: 17897937
- DOI: https://doi.org/10.1074/jbc.M705655200
- Primary Citation of Related Structures:
2QHO - PubMed Abstract:
EDD (or HYD) is an E3 ubiquitin ligase in the family of HECT (homologous to E6-AP C terminus) ligases. EDD contains an N-terminal ubiquitin-associated (UBA) domain, which is present in a variety of proteins involved in ubiquitin-mediated processes. Here, we use isothermal titration calorimetry (ITC), NMR titrations, and pull-down assays to show that the EDD UBA domain binds ubiquitin. The 1.85 A crystal structure of the complex with ubiquitin reveals the structural basis of ubiquitin recognition by UBA helices alpha1 and alpha3. The structure shows a larger number of intermolecular hydrogen bonds than observed in previous UBA/ubiquitin complexes. Two of these involve ordered water molecules. The functional importance of residues at the UBA/ubiquitin interface was confirmed using site-directed mutagenesis. Surface plasmon resonance (SPR) measurements show that the EDD UBA domain does not have a strong preference for polyubiquitin chains over monoubiquitin. This suggests that EDD binds to monoubiquitinated proteins, which is consistent with its involvement in DNA damage repair pathways.
Organizational Affiliation:
Department of Biochemistry, McGill University, Montréal, Québec H3G 1Y6, Canada.