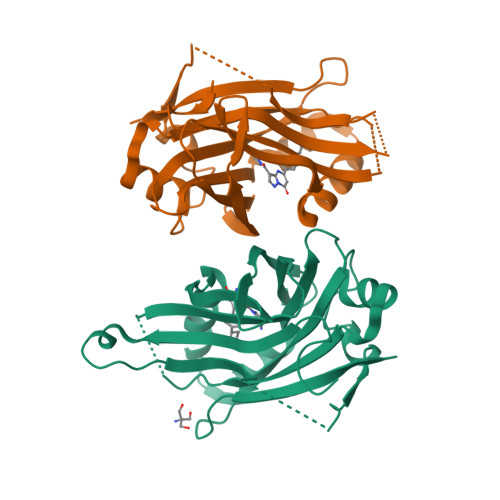
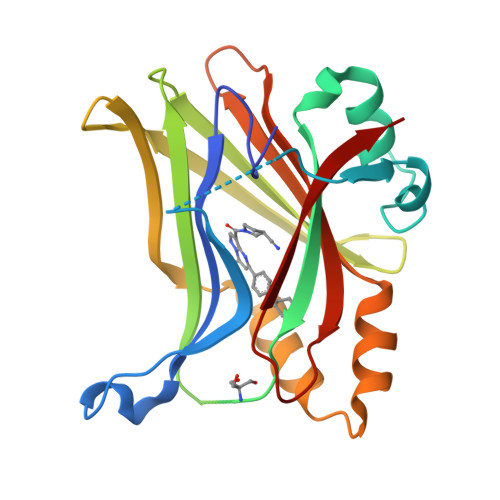
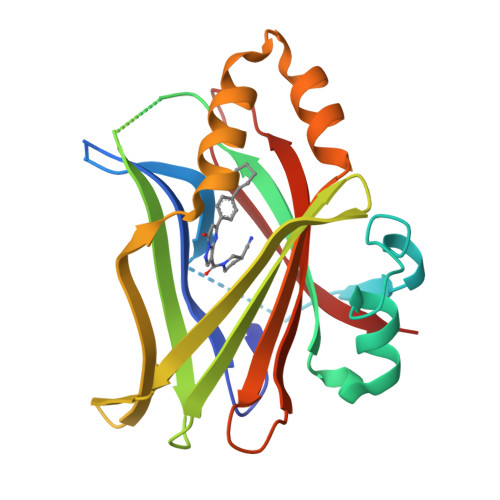
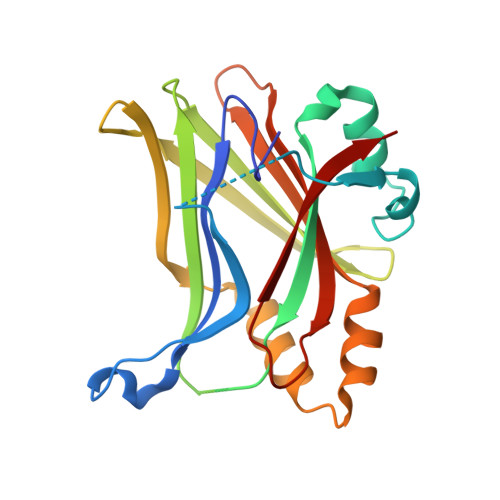
Novel mechanism of YAP-TEAD inhibition results in targeted chromatin remodeling and reveals an expanded Hippo dependent landscape in cancers
Hagenbeek, T.J., Zbieg, J.R., Noland, C.L., Yao, X., Schmidt, S., Clausen, S., Steffek, M., Lee, W., Beroza, P., Martin, S., Lin, E., Fong, R., Di Lello, P., Kubala, M.H., Lacap, J.A., Yang, M.N.Y., Maddalo, D., Lau, J.T., An, L., Levy, E., Lorenzo, M.N., Lee, H.J., Pham, T.H., Modrusan, Z., Zang, R., Chen, Y.C., Li, J., Hafner, M., Chang, M.T., Crawford, J.J., Dey, A.To be published.