Optimization of a Novel DEL Hit That Binds in the Cbl-b SH2 Domain and Blocks Substrate Binding.
Liang, J., Lambrecht, M.J., Arenzana, T.L., Aubert-Nicol, S., Bao, L., Broccatelli, F., Cai, J., Eidenschenk, C., Everett, C., Garner, T., Gruber, F., Haghshenas, P., Huestis, M.P., Hsu, P.L., Kou, P., Jakalian, A., Larouche-Gauthier, R., Leclerc, J.P., Leung, D.H., Martin, A., Murray, J., Prangley, M., Rutz, S., Kakiuchi-Kiyota, S., Satz, A.L., Skelton, N.J., Steffek, M., Stoffler, D., Sudhamsu, J., Tan, S., Wang, J., Wang, S., Wang, Q., Wendorff, T.J., Wichert, M., Yadav, A., Yu, C., Wang, X.(2024) ACS Med Chem Lett 15: 864-872
- PubMed: 38894924
- DOI: https://doi.org/10.1021/acsmedchemlett.4c00068
- Primary Citation of Related Structures:
8VW4, 8VW5 - PubMed Abstract:
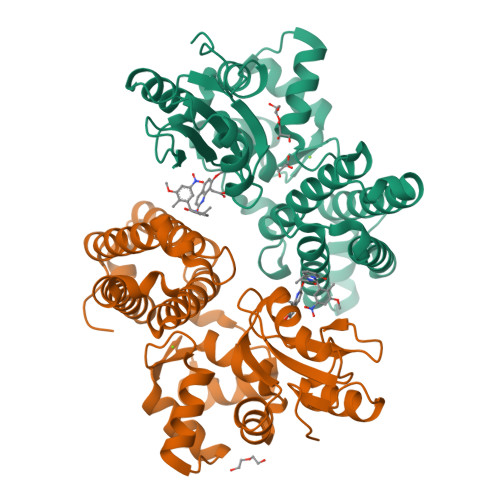
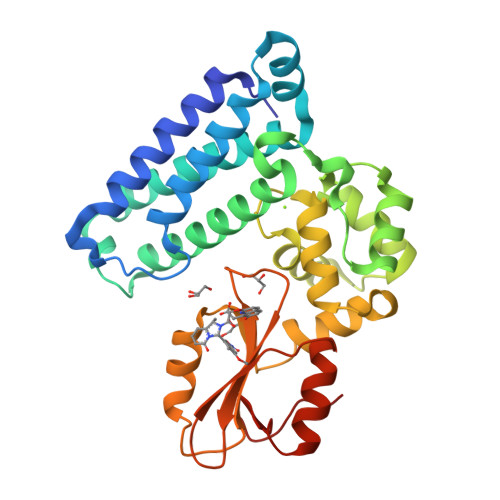
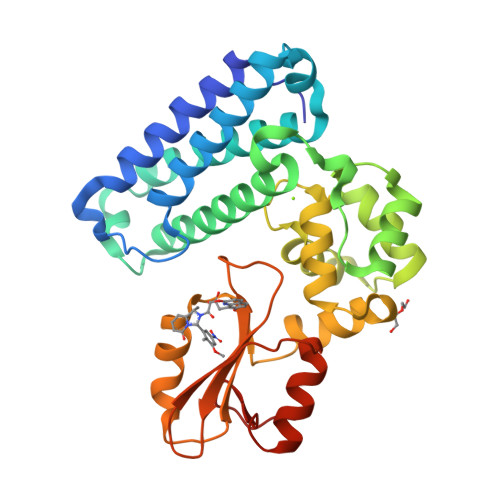
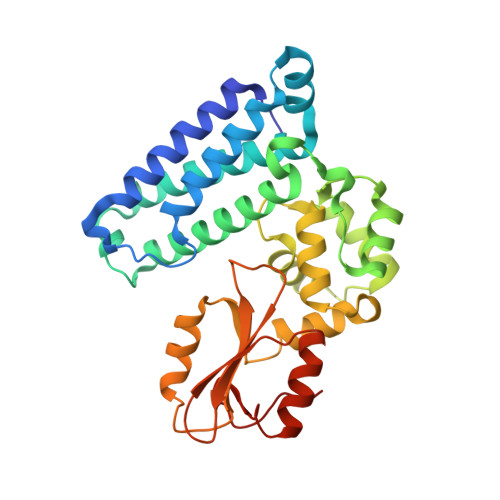
We were attracted to the therapeutic potential of inhibiting Casitas B-lineage lymphoma proto-oncogene-b (Cbl-b), a RING E3 ligase that plays a critical role in regulating the activation of T cells. However, given that only protein-protein interactions were involved, it was unclear whether inhibition by a small molecule would be a viable approach. After screening an ∼6 billion member DNA-encoded library (DEL) using activated Cbl-b, we identified compound 1 as a hit for which the cis -isomer ( 2 ) was confirmed by biochemical and surface plasmon resonance (SPR) assays. Our hit optimization effort was greatly accelerated when we obtained a cocrystal structure of 2 with Cbl-b, which demonstrated induced binding at the substrate binding site, namely, the Src homology-2 (SH2) domain. This was quite noteworthy given that there are few reports of small molecule inhibitors that bind to SH2 domains and block protein-protein interactions. Structure- and property-guided optimization led to compound 27 , which demonstrated measurable cell activity, albeit only at high concentrations.
Organizational Affiliation:
Genentech, Inc., 1 DNA Way, South San Francisco, California 94080, United States.