Full Text |
QUERY: Gene Name = "ICAM1" | MyPDB Login | Search API |
| Search Summary | This query matches 7 Structures. |
Structure Determination MethodologyScientific Name of Source OrganismTaxonomyExperimental MethodPolymer Entity TypeRefinement Resolution (Å)Release DateEnzyme Classification NameSymmetry TypeSCOP Classification | 1 to 7 of 7 Structures Page 1 of 1 Sort by
The Crystal Structure of ICAM-1 D3-D5 fragmentYang, Y., Jun, C.D., Liu, J.H., Zhang, R., Jochimiak, A., Springer, T.A., Wang, J.H. (2004) Mol Cell 14: 269-276
Structural Plasticity in IgSF Domain 4 of ICAM-1 Mediates Cell Surface DimerizationChen, X., Kim, T.D., Carman, C.V., Mi, L., Song, G., Springer, T.A. (2007) Proc Natl Acad Sci U S A 104: 15358-15363
Structure of Engineered Single Domain ICAM-1 D1 with High-Affinity aL Integrin I Domain of Native C-Terminal Helix ConformationKang, S., Kim, C.U., Gu, X., Owens, R.M., van Rijn, S.J., Boonyaleepun, V., Mao, Y., Springer, T.A., Jin, M.M. To be published
The DBLb domain of PF11_0521 PfEMP1 bound to human ICAM-1(2017) Cell Host Microbe 21: 403-414
Coxsackievirus A24v in complex with the D1-D2 fragment of ICAM-1(2018) Proc Natl Acad Sci U S A 115: 397-402
Structure of the PfEMP1 IT4var13 DBLbeta domain bound to ICAM-1(2019) Proc Natl Acad Sci U S A 116: 20124-20134
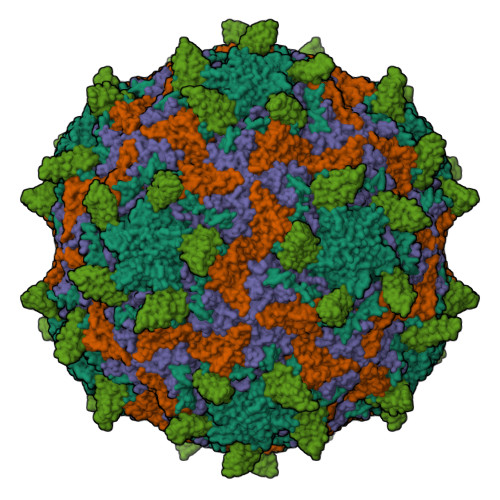
HRV14 in complex with its receptor ICAM-1Hrebik, D., Fuzik, T., Plevka, P. (2021) Proc Natl Acad Sci U S A 118:
1 to 7 of 7 Structures Page 1 of 1 Sort by |