High Resolution X-Ray Structure of an Early Intermediate in the Bacteriorhodopsin Photocycle
Edman, K., Nollert, P., Royant, A., Belrhali, H., Pebay-Peyroula, E., Hajdu, J., Neutze, R., Landau, E.M.(1999) Nature 401: 822
- PubMed: 10548112
- DOI: https://doi.org/10.1038/44623
- Primary Citation of Related Structures:
1QKO, 1QKP - PubMed Abstract:
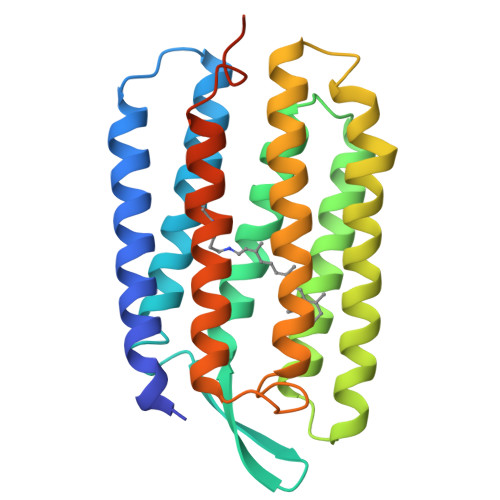
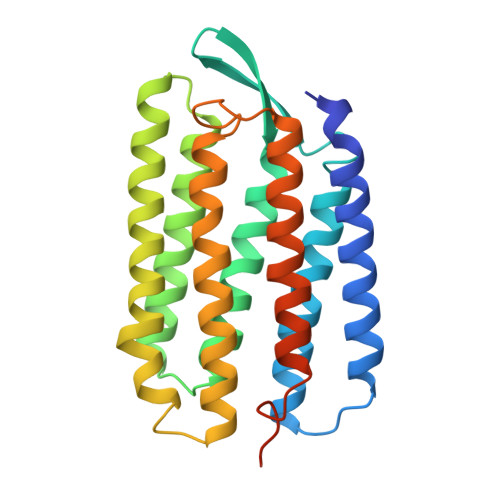
Bacteriorhodopsin is the simplest known photon-driven proton pump and as such provides a model for the study of a basic function in bioenergetics. Its seven transmembrane helices encompass a proton translocation pathway containing the chromophore, a retinal molecule covalently bound to lysine 216 through a protonated Schiff base, and a series of proton donors and acceptors. Photoisomerization of the all-trans retinal to the 13-cis configuration initiates the vectorial translocation of a proton from the Schiff base, the primary proton donor, to the extracellular side, followed by reprotonation of the Schiff base from the cytoplasm. Here we describe the high-resolution X-ray structure of an early intermediate in the photocycle of bacteriorhodopsin, which is formed directly after photoexcitation. A key water molecule is dislocated, allowing the primary proton acceptor, Asp 85, to move. Movement of the main-chain Lys 216 locally disrupts the hydrogen-bonding network of helix G, facilitating structural changes later in the photocycle.
Organizational Affiliation:
Department of Biochemistry, Uppsala University, Biomedical Centre, Sweden.