A redox-controlled molecular switch revealed by the crystal structure of a bacterial heme PAS sensor.
Kurokawa, H., Lee, D.S., Watanabe, M., Sagami, I., Mikami, B., Raman, C.S., Shimizu, T.(2004) J Biological Chem 279: 20186-20193
- PubMed: 14982921
- DOI: https://doi.org/10.1074/jbc.M314199200
- Primary Citation of Related Structures:
1V9Y, 1V9Z - PubMed Abstract:
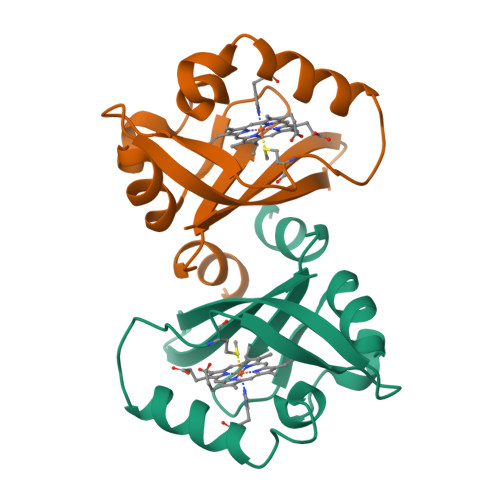
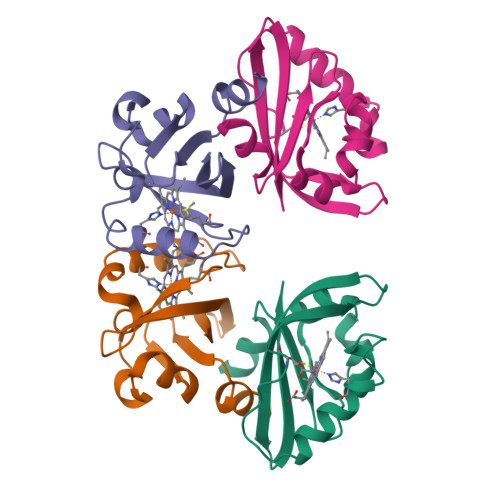
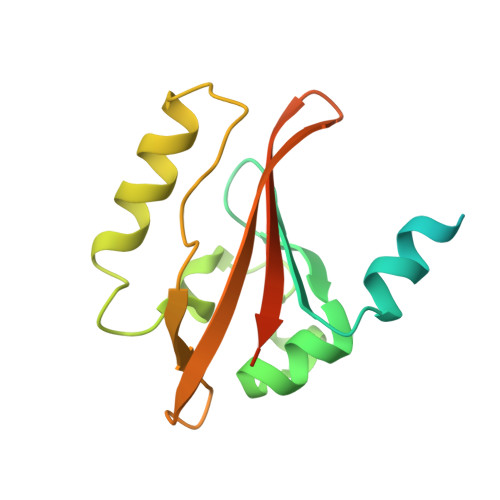
PAS domains, which have been identified in over 1100 proteins from all three kingdoms of life, convert various input stimuli into signals that propagate to downstream components by modifying protein-protein interactions. One such protein is the Escherichia coli redox sensor, Ec DOS, a phosphodiesterase that degrades cyclic adenosine monophosphate in a redox-dependent manner. Here we report the crystal structures of the heme PAS domain of Ec DOS in both inactive Fe(3+) and active Fe(2+) forms at 1.32 and 1.9 A resolution, respectively. The protein folds into a characteristic PAS domain structure and forms a homodimer. In the Fe(3+) form, the heme iron is ligated to a His-77 side chain and a water molecule. Heme iron reduction is accompanied by heme-ligand switching from the water molecule to a side chain of Met-95 from the FG loop. Concomitantly, the flexible FG loop is significantly rigidified, along with a change in the hydrogen bonding pattern and rotation of subunits relative to each other. The present data led us to propose a novel redox-regulated molecular switch in which local heme-ligand switching may trigger a global "scissor-type" subunit movement that facilitates catalytic control.
Organizational Affiliation:
Institute of Multidisciplinary Research for Advanced Materials, Tohoku University, 2-1-1 Katahira, Aoba-ku, Sendai, Miyagi 980-8577, Japan. kurokawa@tagen.tohoku.ac.jp