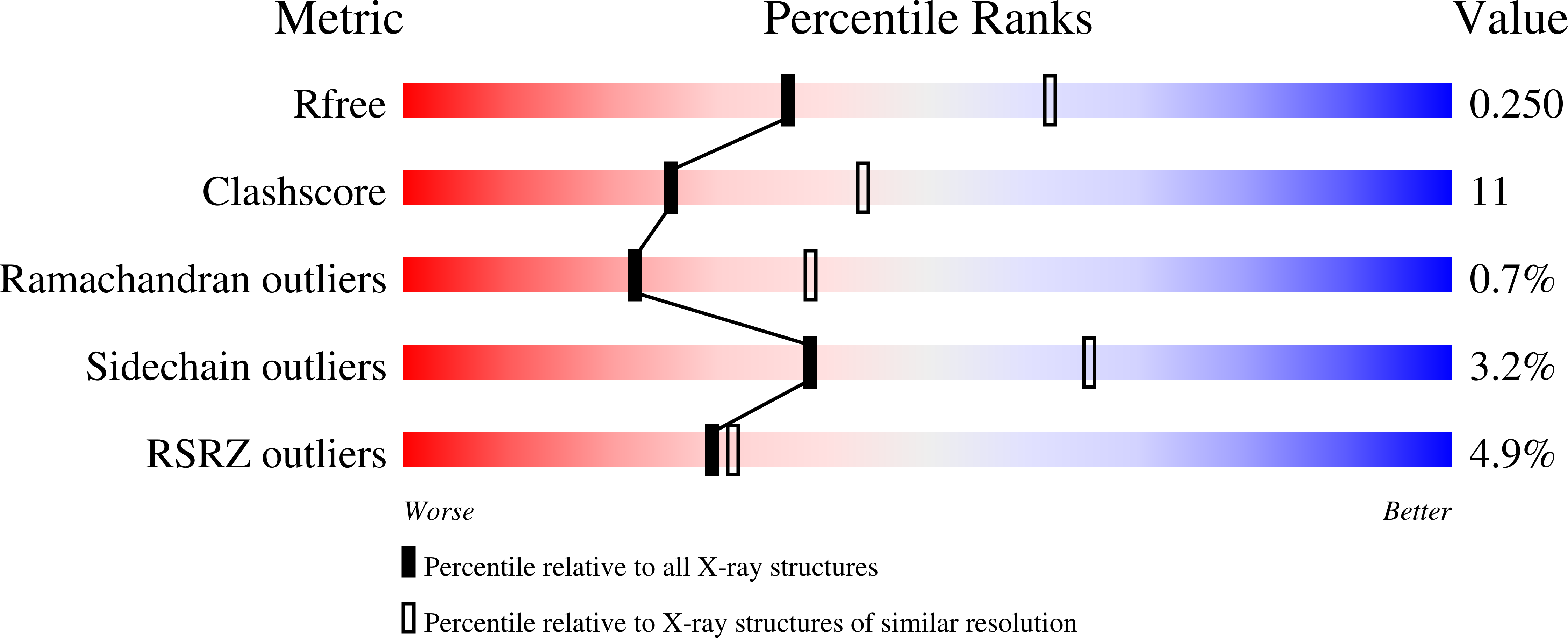
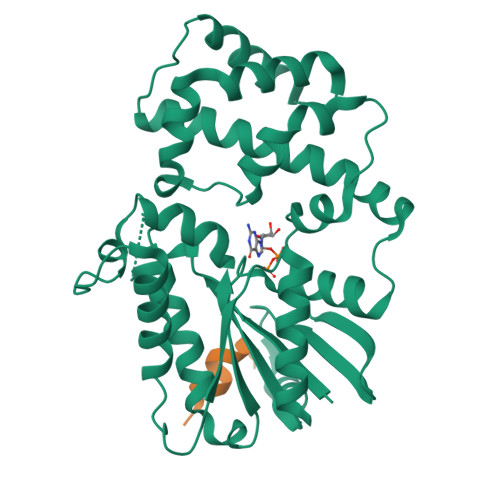
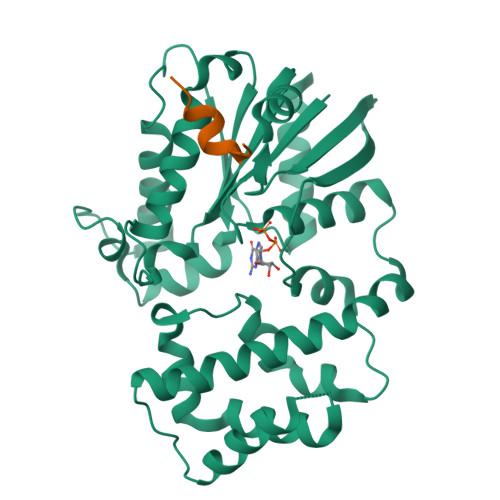
Structure of G-Alpha-I1 bound to a GDP-selective peptide provides insight into guanine nucleotide exchange
Johnston, C.A., Willard, F.S., Jezyk, M.R., Fredericks, Z., Bodor, E.T., Jones, M.B., Blaesius, R., Watts, V.J., Harden, T.K., Sondek, J., Ramer, J.K., Siderovski, D.P.(2005) Structure 7: 1069-1080
- PubMed: 16004878
- DOI: https://doi.org/10.1016/j.str.2005.04.007
- Primary Citation of Related Structures:
1Y3A - PubMed Abstract:
Heterotrimeric G proteins are molecular switches that regulate numerous signaling pathways involved in cellular physiology. This characteristic is achieved by the adoption of two principal states: an inactive, GDP bound state and an active, GTP bound state. Under basal conditions, G proteins exist in the inactive, GDP bound state; thus, nucleotide exchange is crucial to the onset of signaling. Despite our understanding of G protein signaling pathways, the mechanism of nucleotide exchange remains elusive. We employed phage display technology to identify nucleotide state-dependent Galpha binding peptides. Herein, we report a GDP-selective Galpha binding peptide, KB-752, that enhances spontaneous nucleotide exchange of Galpha(i) subunits. Structural determination of the Galpha(i1)/peptide complex reveals unique changes in the Galpha switch regions predicted to enhance nucleotide exchange by creating a GDP dissociation route. Our results cast light onto a potential mechanism by which Galpha subunits adopt a conformation suitable for nucleotide exchange.
Organizational Affiliation:
Department of Pharmacology, The University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.