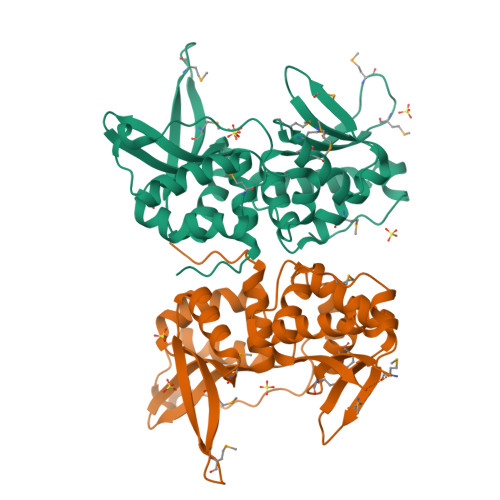
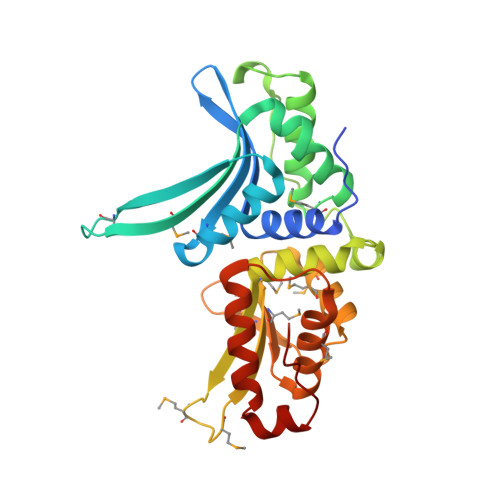
Evolution from DNA to RNA recognition by the bI3 LAGLIDADG maturase
Longo, A., Leonard, C.W., Bassi, G.S., Berndt, D., Krahn, J.M., Hall, T.M., Weeks, K.M.(2005) Nat Struct Mol Biol 12: 779-787
- PubMed: 16116439
- DOI: https://doi.org/10.1038/nsmb976
- Primary Citation of Related Structures:
2AB5 - PubMed Abstract:
LAGLIDADG endonucleases bind across adjacent major grooves via a saddle-shaped surface and catalyze DNA cleavage. Some LAGLIDADG proteins, called maturases, facilitate splicing by group I introns, raising the issue of how a DNA-binding protein and an RNA have evolved to function together. In this report, crystallographic analysis shows that the global architecture of the bI3 maturase is unchanged from its DNA-binding homologs; in contrast, the endonuclease active site, dispensable for splicing facilitation, is efficiently compromised by a lysine residue replacing essential catalytic groups. Biochemical experiments show that the maturase binds a peripheral RNA domain 50 A from the splicing active site, exemplifying long-distance structural communication in a ribonucleoprotein complex. The bI3 maturase nucleic acid recognition saddle interacts at the RNA minor groove; thus, evolution from DNA to RNA function has been mediated by a switch from major to minor groove interaction.
Organizational Affiliation:
Department of Chemistry, University of North Carolina, Chapel Hill, North Carolina 27599-3290, USA.