The Crystal Structure of the SV40 T-Antigen Origin Binding Domain in Complex with DNA
Meinke, G., Phelan, P., Moine, S., Bochkareva, E., Bochkarev, A., Bullock, P.A., Bohm, A.(2007) PLoS Biol 5: e23-e23
- PubMed: 17253903
- DOI: https://doi.org/10.1371/journal.pbio.0050023
- Primary Citation of Related Structures:
2IF9, 2NTC - PubMed Abstract:
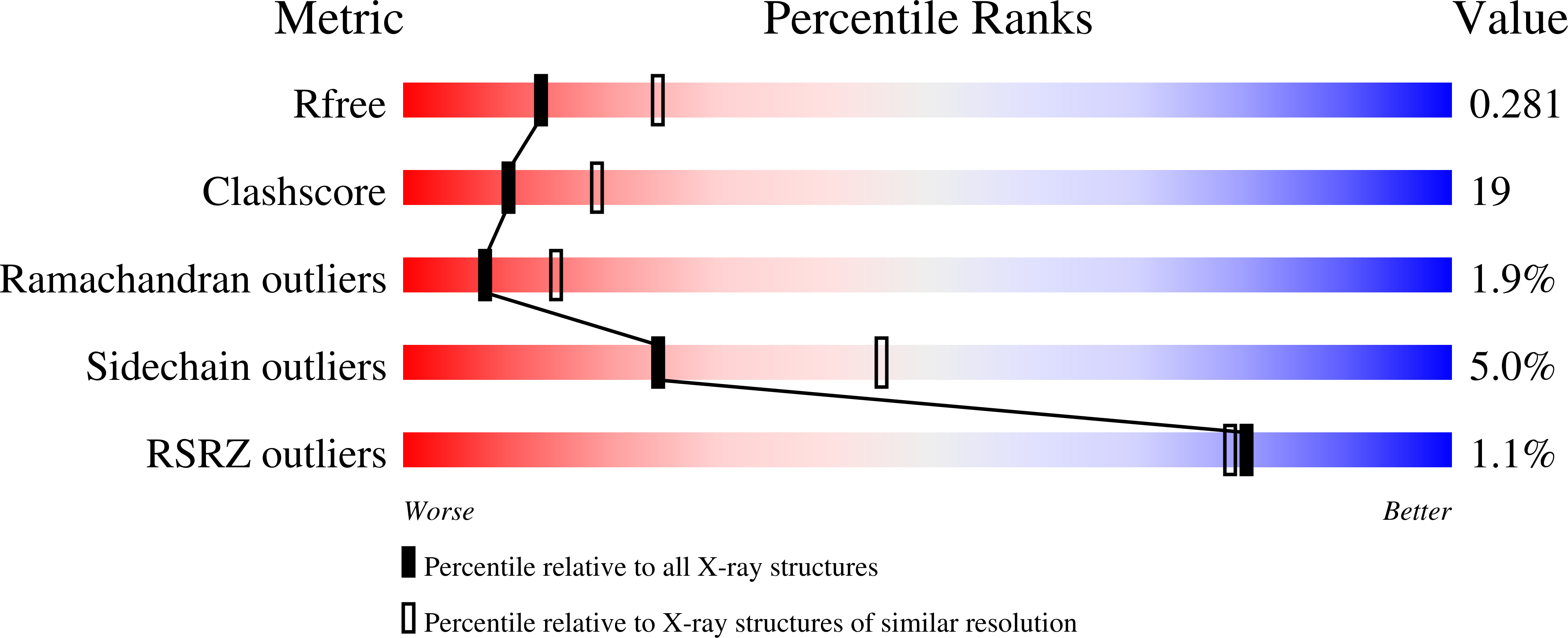
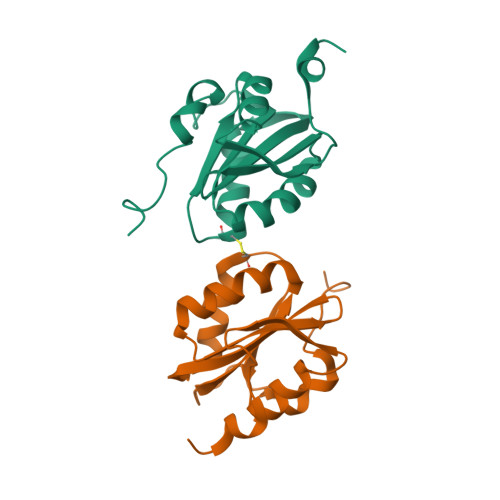
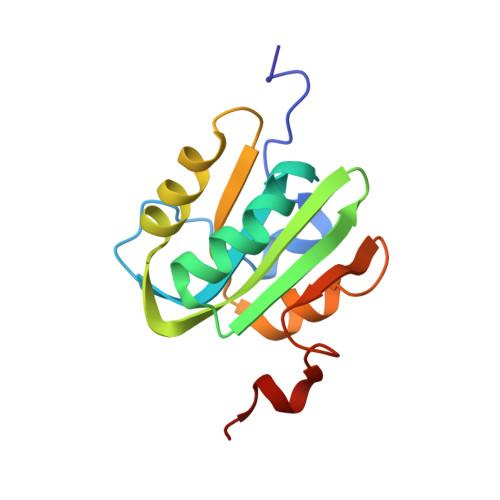
DNA replication is initiated upon binding of "initiators" to origins of replication. In simian virus 40 (SV40), the core origin contains four pentanucleotide binding sites organized as pairs of inverted repeats. Here we describe the crystal structures of the origin binding domain (obd) of the SV40 large T-antigen (T-ag) both with and without a subfragment of origin-containing DNA. In the co-structure, two T-ag obds are oriented in a head-to-head fashion on the same face of the DNA, and each T-ag obd engages the major groove. Although the obds are very close to each other when bound to this DNA target, they do not contact one another. These data provide a high-resolution structural model that explains site-specific binding to the origin and suggests how these interactions help direct the oligomerization events that culminate in assembly of the helicase-active dodecameric complex of T-ag.
Organizational Affiliation:
Department of Biochemistry, School of Medicine, and the Sackler School of Graduate Biomedical Sciences, Tufts University, Boston, Massachusetts, USA.