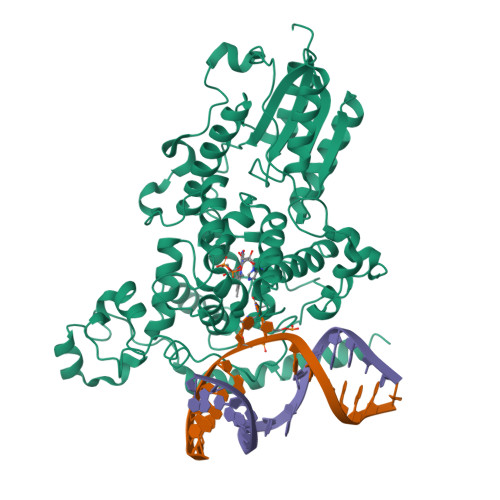
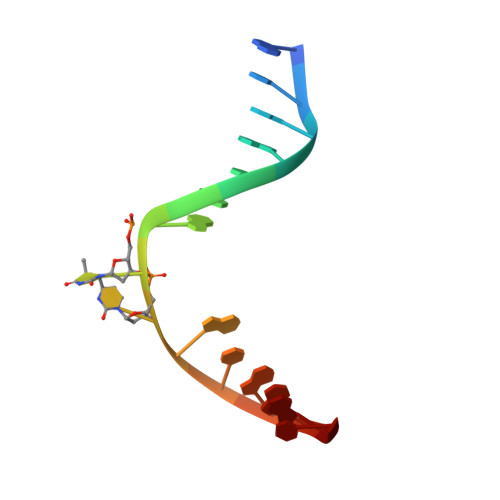
DNA (6-4) Photolyases Reduce Dewar Isomers for Isomerization Into (6-4) Lesions.
Glas, A.F., Kaya, E., Schneider, S., Heil, K., Fazio, D., Maul, M.J., Carell, T.(2010) J Am Chem Soc 132: 3254
- PubMed: 20166732
- DOI: https://doi.org/10.1021/ja910917f
- Primary Citation of Related Structures:
2WQ6, 2WQ7 - PubMed Abstract:
Repair of the Dewar valence isomers by (6-4) photolyases proceeds via an enzyme catalyzed ring-opening reaction of the Dewar lesion to the (6-4) photoproduct.
Organizational Affiliation:
Center for Integrative Protein Science, Department of Chemistry and Biochemistry, Ludwig-Maximilians University Munich, Butenandtstr. 5-13, 81377 Munich, Germany.