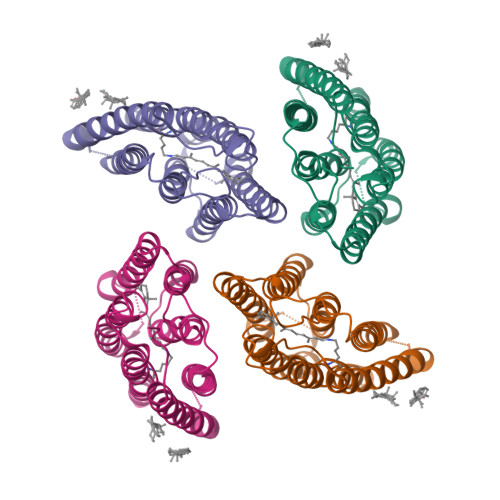
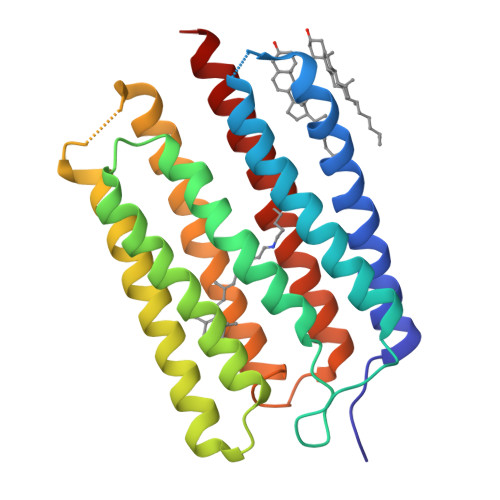
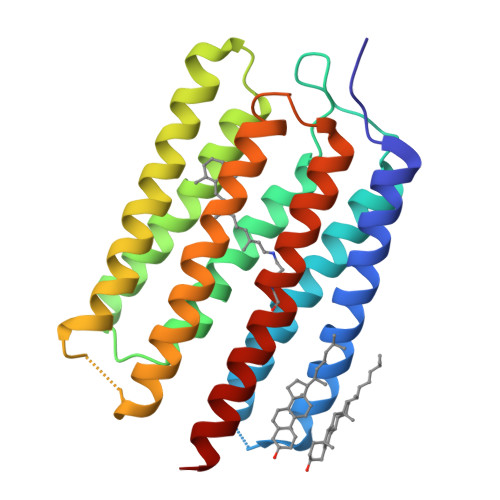
Crystal structure of the eukaryotic light-driven proton-pumping rhodopsin, Acetabularia rhodopsin II, from marine alga
Wada, T., Shimono, K., Kikukawa, T., Hato, M., Shinya, N., Kim, S.Y., Kimura-Someya, T., Shirouzu, M., Tamogami, J., Miyauchi, S., Jung, K.-H., Kamo, N., Yokoyama, S.(2011) J Mol Biology 411: 986-998
- PubMed: 21726566
- DOI: https://doi.org/10.1016/j.jmb.2011.06.028
- Primary Citation of Related Structures:
3AM6 - PubMed Abstract:
Acetabularia rhodopsin (AR) is a rhodopsin from the marine plant Acetabularia acetabulum. The opsin-encoding gene from A. acetabulum, ARII, was cloned and found to be novel but homologous to that reported previously. ARII is a light-driven proton pump, as demonstrated by the existence of a photo-induced current through Xenopus oocytes expressing ARII. The photochemical reaction of ARII prepared by cell-free protein synthesis was similar to that of bacteriorhodopsin (BR), except for the lack of light-dark adaptation and the different proton release and uptake sequence. The crystal structure determined at 3.2 Å resolution is the first structure of a eukaryotic member of the microbial rhodopsin family. The structure of ARII is similar to that of BR. From the cytoplasmic side to the extracellular side of the proton transfer pathway in ARII, Asp92, a Schiff base, Asp207, Asp81, Arg78, Glu199, and Ser189 are arranged in positions similar to those of the corresponding residues directly involved in proton transfer by BR. The side-chain carboxyl group of Asp92 appears to interact with the sulfhydryl group of Cys218, which is unique to ARII and corresponds to Leu223 of BR and to Asp217 of Anabaena sensory rhodopsin. The orientation of the Arg78 side chain is opposite to the corresponding Arg82 of BR. The putative absence of water molecules around Glu199 and Arg78 may disrupt the formation of the low-barrier hydrogen bond at Glu199, resulting in the "late proton release".
Organizational Affiliation:
RIKEN Systems and Structural Biology Center, Yokohama 230-0045, Japan.