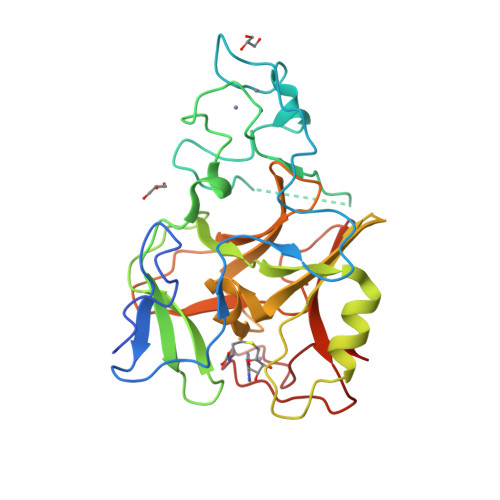
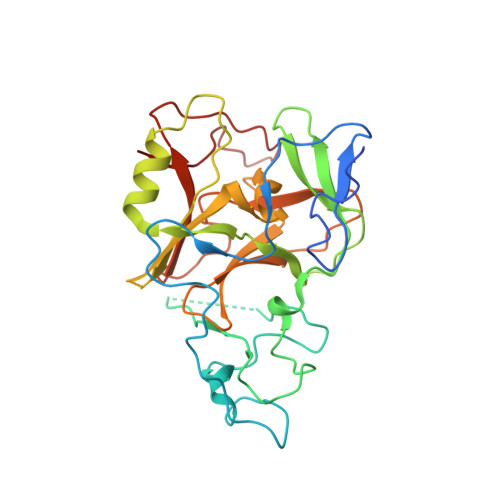
The Crystal Structure of Methyltransferase Domain of Human Histone-lysine N-methyltransferase SETMAR in Complex With AdoHcy.
Wu, H., Lunin, V.V., Ren, H., Dobrovetsky, E., Weigelt, J., Arrowsmith, C.H., Edwards, A.M., Bochkarev, A., Min, J., Plotnikov, A.N.To be published.