Discovery of Cyclic Acylguanidines as Highly Potent and Selective beta-Site Amyloid Cleaving Enzyme (BACE) Inhibitors: Part I-Inhibitor Design and Validation
Zhu, Z., Sun, Z.Y., Ye, Y., Voigt, J., Strickland, C., Smith, E.M., Cumming, J., Wang, L., Wong, J., Wang, Y.S., Wyss, D.F., Chen, X., Kuvelkar, R., Kennedy, M.E., Favreau, L., Parker, E., McKittrick, B.A., Stamford, A., Czarniecki, M., Greenlee, W., Hunter, J.C.(2010) J Med Chem 53: 951-965
- PubMed: 20043696
- DOI: https://doi.org/10.1021/jm901408p
- Primary Citation of Related Structures:
3L58, 3L59, 3L5B, 3L5C, 3L5D, 3L5E, 3L5F - PubMed Abstract:
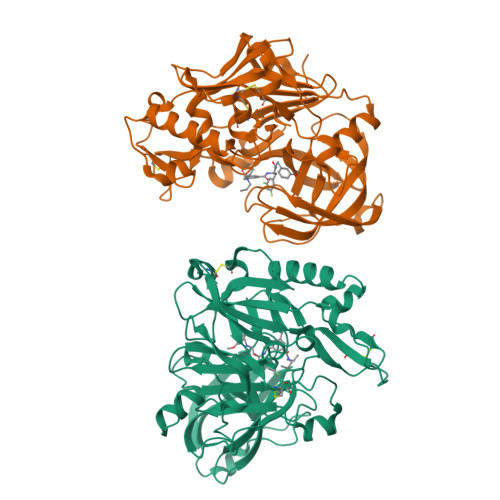
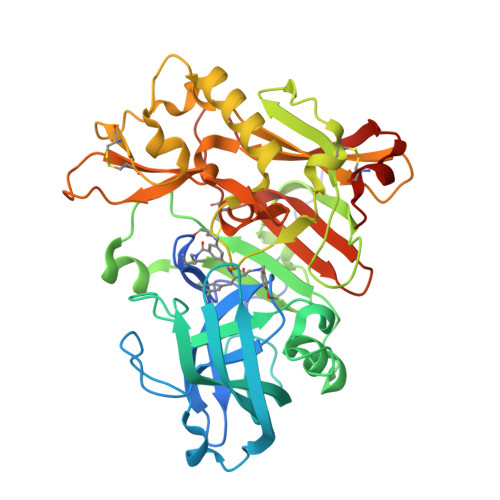
A number of novel amidine containing heterocycles were designed to reproduce the unique interaction pattern, revealed by X-ray crystallography, between the BACE-1 catalytic diad and a weak NMR screening hit (3), with special attention paid to maintaining the appropriate basicity and limiting the number of H-bonding donors of these scaffolds. The iminohydantoin cores (10 and 23) were examined first and found to interact with the catalytic diad in one of two binding modes (A and B), each with the iminohydantoin core flipped 180 degrees in relation to the other. The amidine structural motif within each core forms a bidentate interaction with a different aspartic acid of the catalytic diad. Both modes reproduced a highly conserved interaction pattern between the inhibitors and the catalytic aspartates, as revealed by 3. Potent iminohydantoin BACE-1 inhibitors have been obtained, validating the molecular design as aspartyl protease catalytic site inhibitors. Brain penetrant small molecule BACE inhibitors with high ligand efficiencies have been discovered, enabling multiple strategies for further development of these inhibitors into highly potent, selective and in vivo efficacious BACE inhibitors.
Organizational Affiliation:
Department of Medicinal Chemistry, Schering-Plough Research Institute, 2015 Galloping Hill Road, Kenilworth, New Jersey 07033, USA. Zhaoning.Zhu@spcorp.com