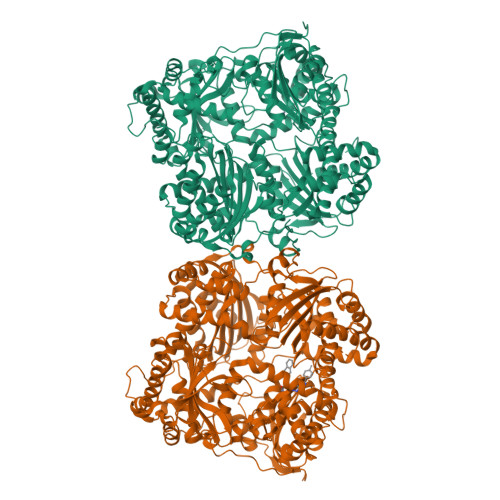
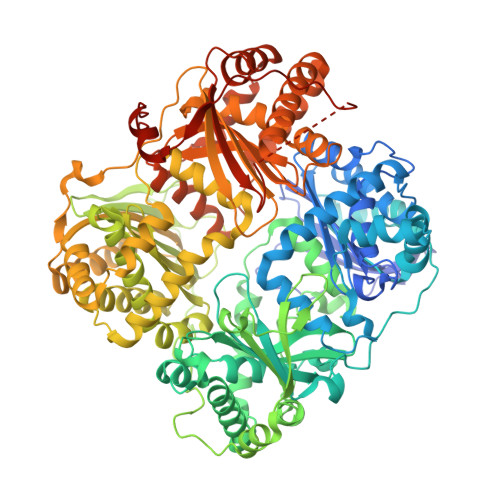
Catalytic site inhibition of insulin-degrading enzyme by a small molecule induces glucose intolerance in mice.
Deprez-Poulain, R., Hennuyer, N., Bosc, D., Liang, W.G., Enee, E., Marechal, X., Charton, J., Totobenazara, J., Berte, G., Jahklal, J., Verdelet, T., Dumont, J., Dassonneville, S., Woitrain, E., Gauriot, M., Paquet, C., Duplan, I., Hermant, P., Cantrelle, F.X., Sevin, E., Culot, M., Landry, V., Herledan, A., Piveteau, C., Lippens, G., Leroux, F., Tang, W.J., van Endert, P., Staels, B., Deprez, B.(2015) Nat Commun 6: 8250-8250
- PubMed: 26394692
- DOI: https://doi.org/10.1038/ncomms9250
- Primary Citation of Related Structures:
4IFH, 4NXO, 4RE9 - PubMed Abstract:
Insulin-degrading enzyme (IDE) is a protease that cleaves insulin and other bioactive peptides such as amyloid-β. Knockout and genetic studies have linked IDE to Alzheimer's disease and type-2 diabetes. As the major insulin-degrading protease, IDE is a candidate drug target in diabetes. Here we have used kinetic target-guided synthesis to design the first catalytic site inhibitor of IDE suitable for in vivo studies (BDM44768). Crystallographic and small angle X-ray scattering analyses show that it locks IDE in a closed conformation. Among a panel of metalloproteases, BDM44768 selectively inhibits IDE. Acute treatment of mice with BDM44768 increases insulin signalling and surprisingly impairs glucose tolerance in an IDE-dependent manner. These results confirm that IDE is involved in pathways that modulate short-term glucose homeostasis, but casts doubt on the general usefulness of the inhibition of IDE catalytic activity to treat diabetes.
Organizational Affiliation:
Institut Pasteur de Lille, Lille F-59000, France.