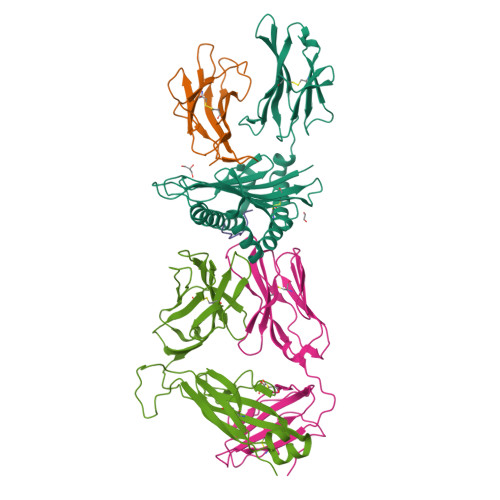
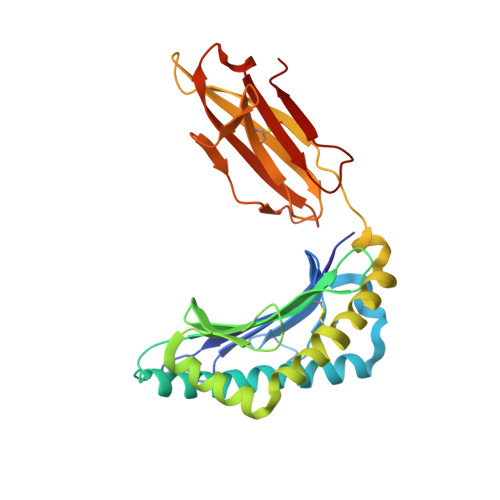
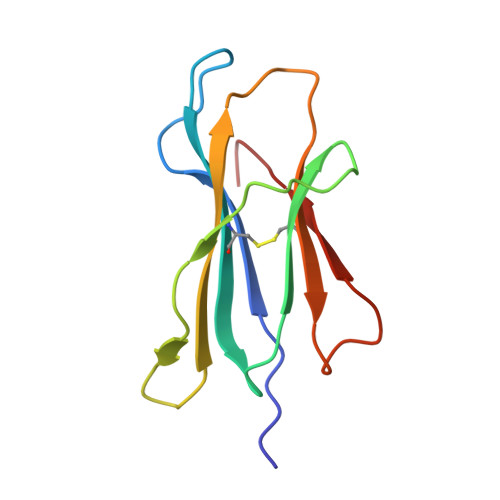
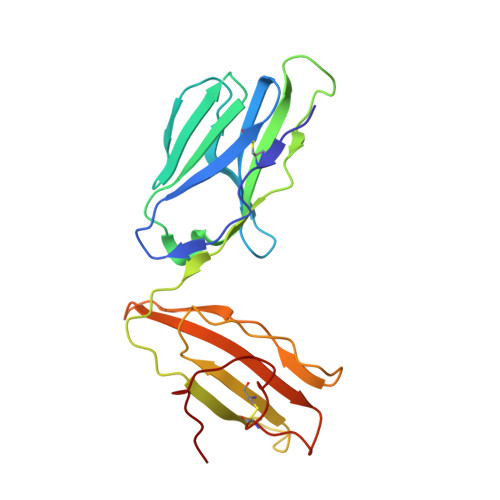
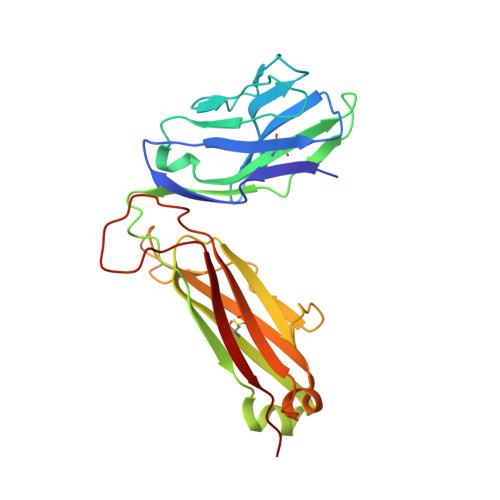
T-cell receptor (TCR)-peptide specificity overrides affinity-enhancing TCR-major histocompatibility complex interactions.
Cole, D.K., Miles, K.M., Madura, F., Holland, C.J., Schauenburg, A.J., Godkin, A.J., Bulek, A.M., Fuller, A., Akpovwa, H.J., Pymm, P.G., Liddy, N., Sami, M., Li, Y., Rizkallah, P.J., Jakobsen, B.K., Sewell, A.K.(2014) J Biological Chem 289: 628-638
- PubMed: 24196962
- DOI: https://doi.org/10.1074/jbc.M113.522110
- Primary Citation of Related Structures:
4MNQ - PubMed Abstract:
αβ T-cell receptors (TCRs) engage antigens using complementarity-determining region (CDR) loops that are either germ line-encoded (CDR1 and CDR2) or somatically rearranged (CDR3). TCR ligands compose a presentation platform (major histocompatibility complex (MHC)) and a variable antigenic component consisting of a short "foreign" peptide. The sequence of events when the TCR engages its peptide-MHC (pMHC) ligand remains unclear. Some studies suggest that the germ line elements of the TCR engage the MHC prior to peptide scanning, but this order of binding is difficult to reconcile with some TCR-pMHC structures. Here, we used TCRs that exhibited enhanced pMHC binding as a result of mutations in either CDR2 and/or CDR3 loops, that bound to the MHC or peptide, respectively, to dissect the roles of these loops in stabilizing TCR-pMHC interactions. Our data show that TCR-peptide interactions play a strongly dominant energetic role providing a binding mode that is both temporally and energetically complementary with a system requiring positive selection by self-pMHC in the thymus and rapid recognition of non-self-pMHC in the periphery.
Organizational Affiliation:
From Cardiff University School of Medicine, Heath Park, Cardiff CF14 4XN.