From bacterial to human dihydrouridine synthase: automated structure determination.
Whelan, F., Jenkins, H.T., Griffiths, S.C., Byrne, R.T., Dodson, E.J., Antson, A.A.(2015) Acta Crystallogr D Biol Crystallogr 71: 1564-1571
- PubMed: 26143927
- DOI: https://doi.org/10.1107/S1399004715009220
- Primary Citation of Related Structures:
4XP7 - PubMed Abstract:
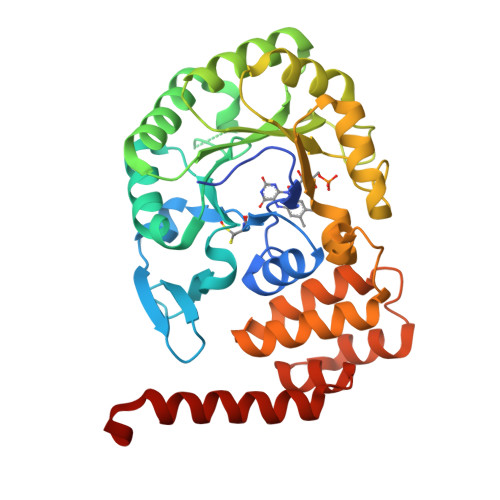
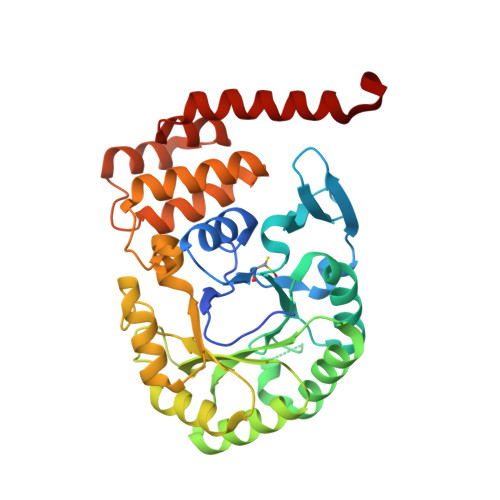
The reduction of uridine to dihydrouridine at specific positions in tRNA is catalysed by dihydrouridine synthase (Dus) enzymes. Increased expression of human dihydrouridine synthase 2 (hDus2) has been linked to pulmonary carcinogenesis, while its knockdown decreased cancer cell line viability, suggesting that it may serve as a valuable target for therapeutic intervention. Here, the X-ray crystal structure of a construct of hDus2 encompassing the catalytic and tRNA-recognition domains (residues 1-340) determined at 1.9 Å resolution is presented. It is shown that the structure can be determined automatically by phenix.mr_rosetta starting from a bacterial Dus enzyme with only 18% sequence identity and a significantly divergent structure. The overall fold of the human Dus2 is similar to that of bacterial enzymes, but has a larger recognition domain and a unique three-stranded antiparallel β-sheet insertion into the catalytic domain that packs next to the recognition domain, contributing to domain-domain interactions. The structure may inform the development of novel therapeutic approaches in the fight against lung cancer.
Organizational Affiliation:
Department of Biology, The University of York, Heslington, York YO10 5DD, England.