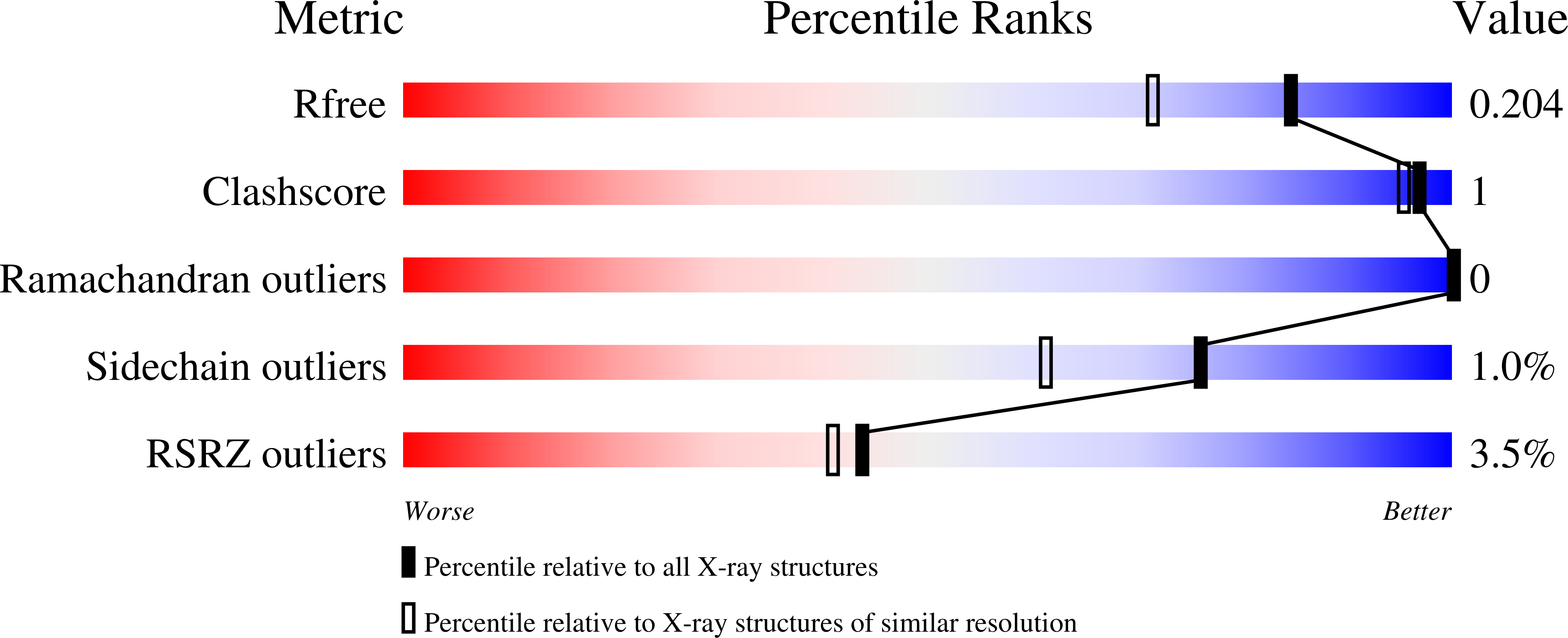
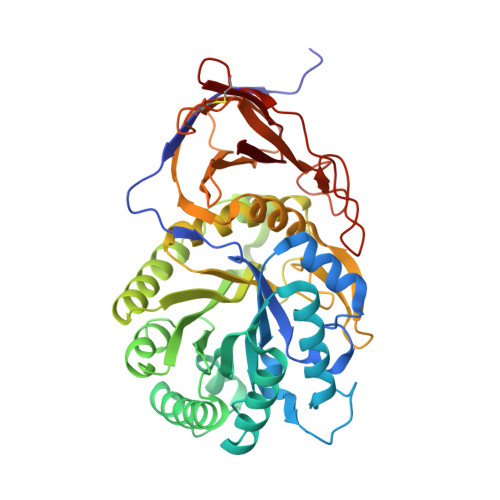
Functional and structural characterization of a heparanase.
Bohlmann, L., Tredwell, G.D., Yu, X., Chang, C.W., Haselhorst, T., Winger, M., Dyason, J.C., Thomson, R.J., Tiralongo, J., Beacham, I.R., Blanchard, H., von Itzstein, M.(2015) Nat Chem Biol 11: 955-957
- PubMed: 26565989
- DOI: https://doi.org/10.1038/nchembio.1956
- Primary Citation of Related Structures:
5BWI - PubMed Abstract:
We report the structural and functional characterization of a novel heparanase (BpHep) from the invasive pathogenic bacterium Burkholderia pseudomallei (Bp), showing ∼24% sequence identity with human heparanase (hHep). Site-directed mutagenesis studies confirmed the active site resi-dues essential for activity, and we found that BpHep has specificity for heparan sulfate. Finally, we describe the first heparanase X-ray crystal structure, which provides new insight into both substrate recognition and inhibitor design.
Organizational Affiliation:
Institute for Glycomics, Griffith University, Gold Coast Campus, Southport, Queensland, Australia.