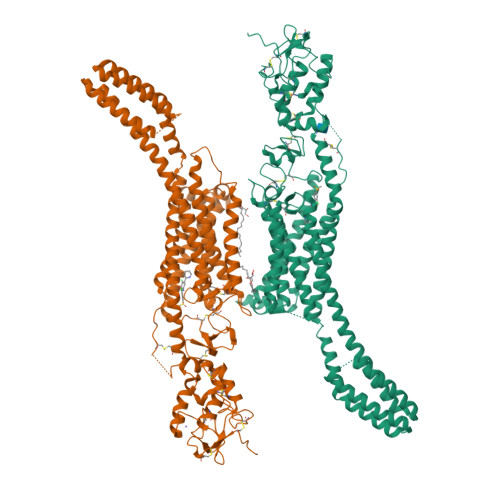
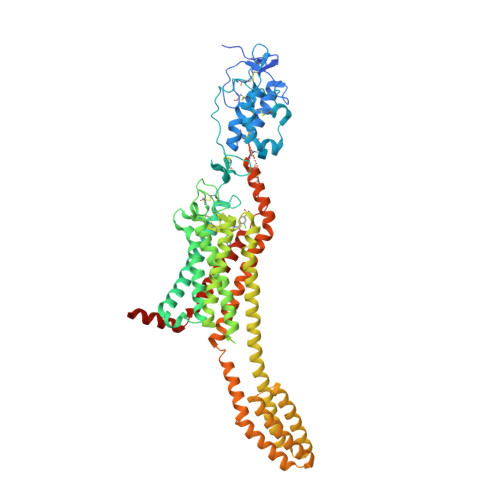
Structural basis of Smoothened regulation by its extracellular domains.
Byrne, E.F., Sircar, R., Miller, P.S., Hedger, G., Luchetti, G., Nachtergaele, S., Tully, M.D., Mydock-McGrane, L., Covey, D.F., Rambo, R.P., Sansom, M.S., Newstead, S., Rohatgi, R., Siebold, C.(2016) Nature 535: 517-522
- PubMed: 27437577
- DOI: https://doi.org/10.1038/nature18934
- Primary Citation of Related Structures:
5L7D, 5L7I - PubMed Abstract:
Developmental signals of the Hedgehog (Hh) and Wnt families are transduced across the membrane by Frizzledclass G-protein-coupled receptors (GPCRs) composed of both a heptahelical transmembrane domain (TMD) and an extracellular cysteine-rich domain (CRD). How the large extracellular domains of GPCRs regulate signalling by the TMD is unknown. We present crystal structures of the Hh signal transducer and oncoprotein Smoothened, a GPCR that contains two distinct ligand-binding sites: one in its TMD and one in the CRD. The CRD is stacked a top the TMD, separated by an intervening wedge-like linker domain. Structure-guided mutations show that the interface between the CRD, linker domain and TMD stabilizes the inactive state of Smoothened. Unexpectedly, we find a cholesterol molecule bound to Smoothened in the CRD binding site. Mutations predicted to prevent cholesterol binding impair the ability of Smoothened to transmit native Hh signals. Binding of a clinically used antagonist, vismodegib, to the TMD induces a conformational change that is propagated to the CRD, resulting in loss of cholesterol from the CRD-linker domain-TMD interface. Our results clarify the structural mechanism by which the activity of a GPCR is controlled by ligand-regulated interactions between its extracellular and transmembrane domains.
Organizational Affiliation:
Division of Structural Biology, Wellcome Trust Centre for Human Genetics, University of Oxford, Oxford, UK.