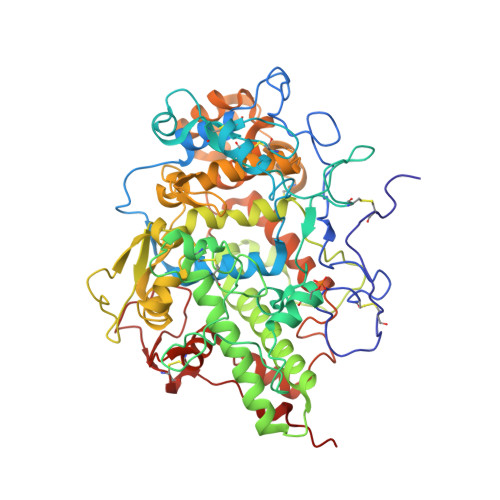
Structural basis of activation of mammalian heme peroxidases
Singh, P.K., Iqbal, N., Sirohi, H.V., Bairagya, H.R., Kaur, P., Sharma, S., Singh, T.P.(2018) Prog Biophys Mol Biol 133: 49-55
- PubMed: 29174286
- DOI: https://doi.org/10.1016/j.pbiomolbio.2017.11.003
- Primary Citation of Related Structures:
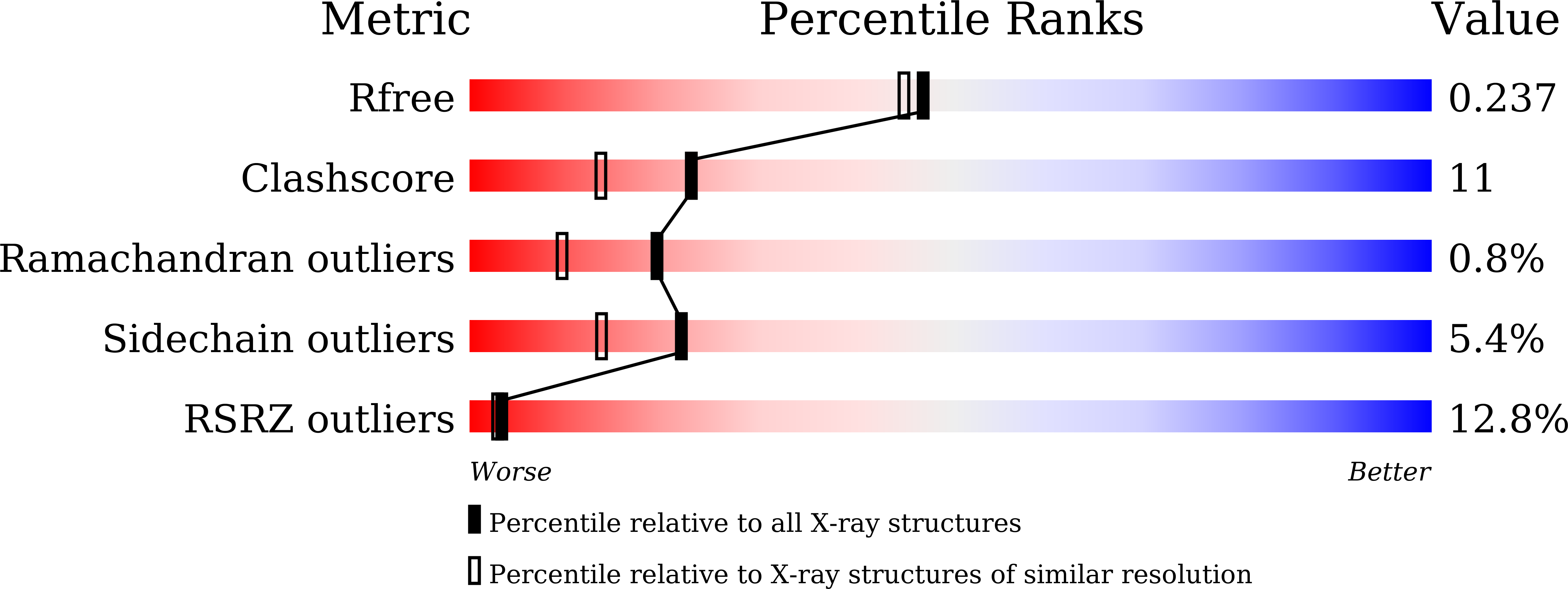
5WV3 - PubMed Abstract:
The mammalian heme peroxidases including lactoperoxidase (LPO), myeloperoxidase (MPO), eosinophil peroxidase (EPO) and thyroid peroxidase (TPO) contain a covalently linked heme moiety. Initially, it was believed that the heme group was fully cross-linked to protein molecule through at least two ester linkages involving conserved glutamate and aspartate residues with 1-methyl and 5-methyl groups of pyrrole rings A and C respectively. In MPO, an additional sulfonium ion linkage was present between 2-vinyl group of pyrrole ring A of the heme moiety and a methionine residue of the protein. These linkages were formed through a self processing mechanism. Subsequently, biochemical studies indicated that the heme moiety was partially attached to protein. The recent structural studies have shown that the covalent linkage involving glutamate and 1-methyl group of pyrrole ring of heme moiety was partially formed. When glutamate is not covalently linked to heme moiety, its side chain occupies a position in the substrate binding site on the distal heme side and blocks the substrate binding site leading to inactivation. However, an exposure to H 2 O 2 converts it to a fully covalently linked state with heme. Thus in mammalian heme peroxidases, the Glu-heme linkage is essential for catalytic action.
Organizational Affiliation:
Department of Biophysics, All India Institute of Medical Sciences, New Delhi, India.