A semi-synthetic strategy for derivatization of the violacein natural product scaffold
Lai, H.E., Chee, S.M., Morgan, M., Moore, S.J., Polizzi, K.M., Freemont, P.S.(2018) bioRxiv
Experimental Data Snapshot
Starting Model: experimental
View more details
(2018) bioRxiv
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Flavin-dependent L-tryptophan oxidase VioA | A [auth B], B [auth A] | 417 | Chromobacterium violaceum ATCC 12472 | Mutation(s): 0 Gene Names: vioA, CV_3274 EC: 1.4.3.23 | 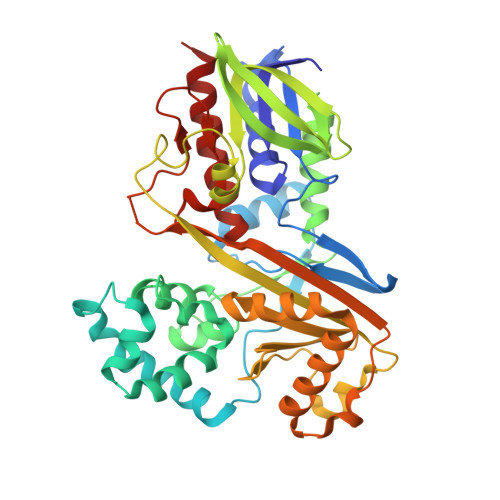 |
UniProt | |||||
Find proteins for Q9S3V1 (Chromobacterium violaceum (strain ATCC 12472 / DSM 30191 / JCM 1249 / CCUG 213 / NBRC 12614 / NCIMB 9131 / NCTC 9757 / MK)) Explore Q9S3V1 Go to UniProtKB: Q9S3V1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9S3V1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| FAD Query on FAD | C [auth B], G [auth A] | FLAVIN-ADENINE DINUCLEOTIDE C27 H33 N9 O15 P2 VWWQXMAJTJZDQX-UYBVJOGSSA-N |  | ||
| TRP Query on TRP | D [auth B], H [auth A] | TRYPTOPHAN C11 H12 N2 O2 QIVBCDIJIAJPQS-VIFPVBQESA-N |  | ||
| EDO Query on EDO | E [auth B], I [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| MG Query on MG | F [auth B], J [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 152.092 | α = 90 |
| b = 175.338 | β = 90 |
| c = 94.436 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| xia2 | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Imperial College London | United Kingdom | President's PhD Scholarship |