Multitarget, Selective Compound Design Yields Potent Inhibitors of a Kinetoplastid Pteridine Reductase 1.
Pohner, I., Quotadamo, A., Panecka-Hofman, J., Luciani, R., Santucci, M., Linciano, P., Landi, G., Di Pisa, F., Dello Iacono, L., Pozzi, C., Mangani, S., Gul, S., Witt, G., Ellinger, B., Kuzikov, M., Santarem, N., Cordeiro-da-Silva, A., Costi, M.P., Venturelli, A., Wade, R.C.(2022) J Med Chem 65: 9011-9033
- PubMed: 35675511
- DOI: https://doi.org/10.1021/acs.jmedchem.2c00232
- Primary Citation of Related Structures:
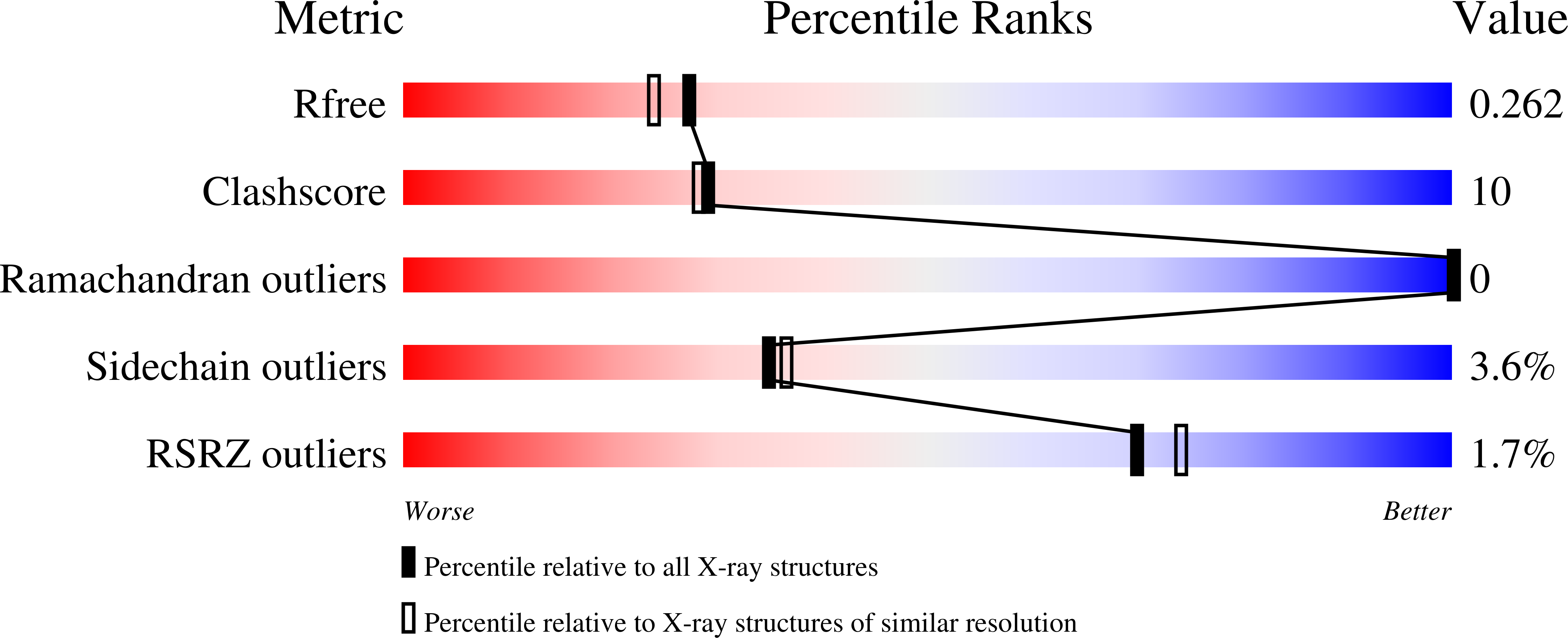
6RX0, 6RX5, 6RX6, 6RXC - PubMed Abstract:
The optimization of compounds with multiple targets is a difficult multidimensional problem in the drug discovery cycle. Here, we present a systematic, multidisciplinary approach to the development of selective antiparasitic compounds. Computational fragment-based design of novel pteridine derivatives along with iterations of crystallographic structure determination allowed for the derivation of a structure-activity relationship for multitarget inhibition. The approach yielded compounds showing apparent picomolar inhibition of T. brucei pteridine reductase 1 (PTR1), nanomolar inhibition of L. major PTR1, and selective submicromolar inhibition of parasite dihydrofolate reductase (DHFR) versus human DHFR. Moreover, by combining design for polypharmacology with a property-based on-parasite optimization, we found three compounds that exhibited micromolar EC 50 values against T. brucei brucei while retaining their target inhibition. Our results provide a basis for the further development of pteridine-based compounds, and we expect our multitarget approach to be generally applicable to the design and optimization of anti-infective agents.
Organizational Affiliation:
Molecular and Cellular Modeling Group, Heidelberg Institute for Theoretical Studies (HITS), D-69118 Heidelberg, Germany.