Novel Selenium-based compounds with therapeutic potential for SOD1-linked amyotrophic lateral sclerosis.
Amporndanai, K., Rogers, M., Watanabe, S., Yamanaka, K., O'Neill, P.M., Hasnain, S.S.(2020) EBioMedicine 59: 102980-102980
- PubMed: 32862101
- DOI: https://doi.org/10.1016/j.ebiom.2020.102980
- Primary Citation of Related Structures:
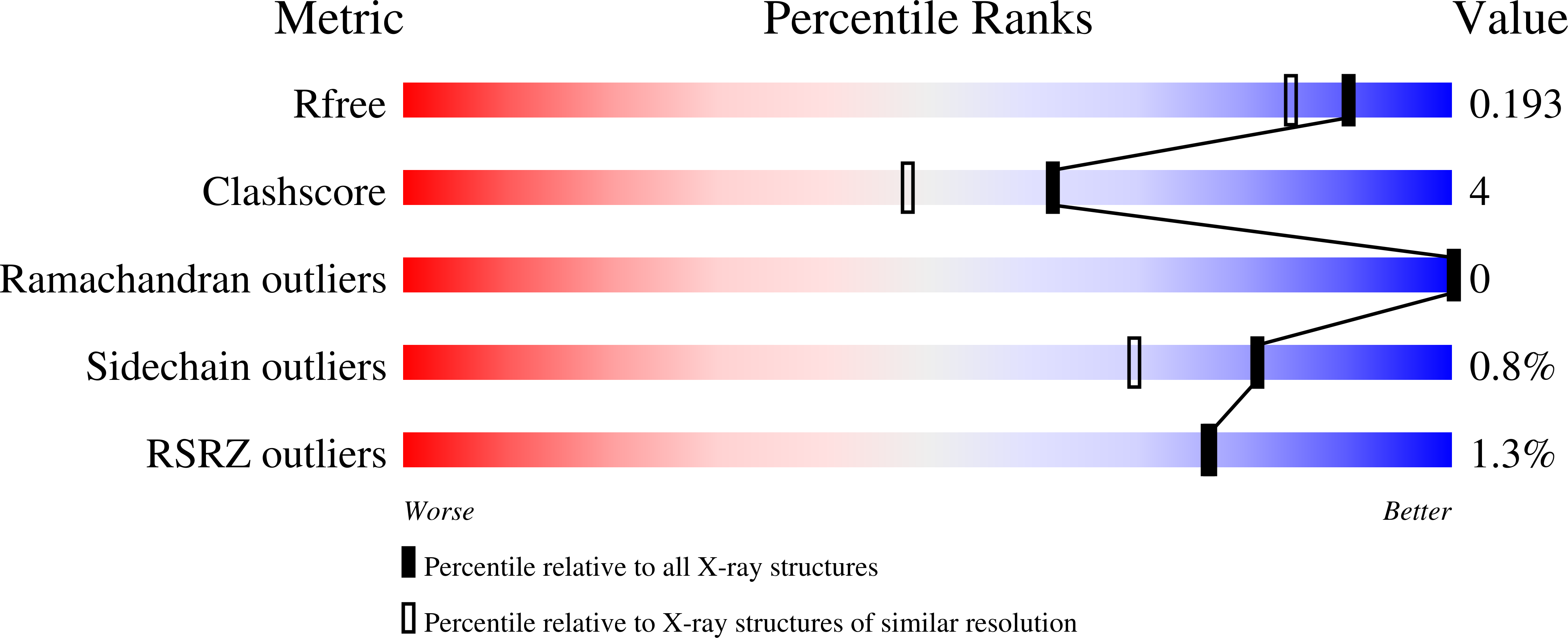
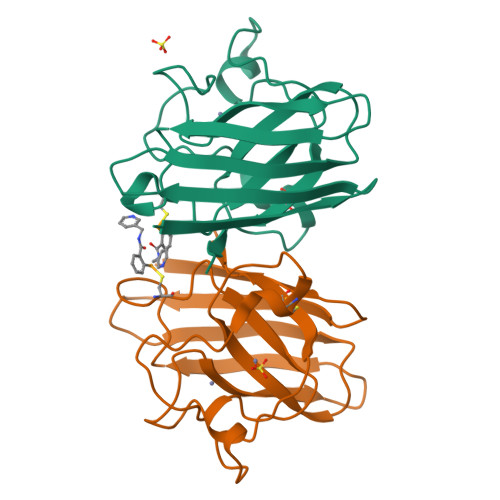
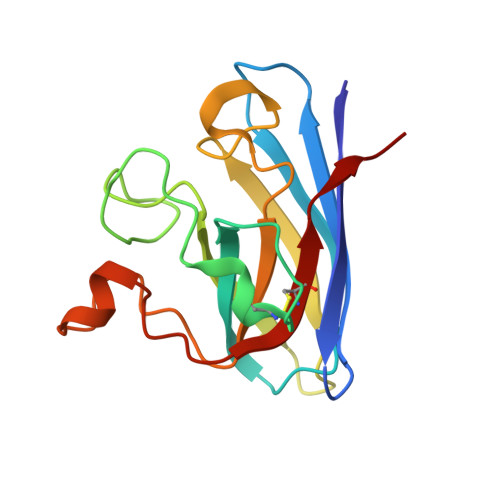
6Z3V, 6Z4G, 6Z4H, 6Z4I, 6Z4J, 6Z4K, 6Z4L, 6Z4M, 6Z4O - PubMed Abstract:
Amyotrophic lateral sclerosis (ALS), also known as motor neuron disease as well as Lou Gehrig's disease, is a progressive neurological disorder selectively affecting motor neurons with no currently known cure. Around 20% of the familial ALS cases arise from dominant mutations in the sod1 gene encoding superoxide dismutase1 (SOD1) enzyme. Aggregation of mutant SOD1 in familial cases and of wild-type SOD1 in at least some sporadic ALS cases is one of the known causes of the disease. Riluzole, approved in 1995 and edaravone in 2017 remain the only drugs with limited therapeutic benefits. We have utilised the ebselen template to develop novel compounds that redeem stability of mutant SOD1 dimer and prevent aggregation. Binding modes of compounds have been visualised by crystallography. In vitro neuroprotection and toxicity of lead compounds have been performed in mouse neuronal cells and disease onset delay of ebselen has been demonstrated in transgenic ALS mice model. We have developed a number of ebselen-based compounds with improvements in A4V SOD1 stabilisation and in vitro therapeutic effects with significantly better potency than edaravone. Structure-activity relationship of hits has been guided by high resolution structures of ligand-bound A4V SOD1. We also show clear disease onset delay of ebselen in transgenic ALS mice model holding encouraging promise for potential therapeutic compounds. Our finding established the new generation of organo-selenium compounds with better in vitro neuroprotective activity than edaravone. The potential of this class of compounds may offer an alternative therapeutic agent for ALS treatment. The ability of these compounds to target cysteine 111 in SOD may have wider therapeutic applications targeting cysteines of enzymes involved in pathogenic and viral diseases including main protease of SARS-Cov-2 (COVID-19). Project funding was supported by the ALS Association grant (WA1128) and Fostering Joint International Research (19KK0214) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
Organizational Affiliation:
Molecular Biophysics Group, Department of Biochemistry and System Biology, Institute of System, Molecular and Integrative Biology, Faculty of Health and Life Sciences, University of Liverpool, Liverpool, L69 7ZB, United Kingdom.