Discovery of a new class of reversible TEA-domain transcription factor inhibitors with a novel binding mode.
Hu, L., Sun, Y., Liu, S., Erb, H., Singh, A., Mao, J., Luo, X., Wu, X.(2022) Elife 11
- PubMed: 36398861
- DOI: https://doi.org/10.7554/eLife.80210
- Primary Citation of Related Structures:
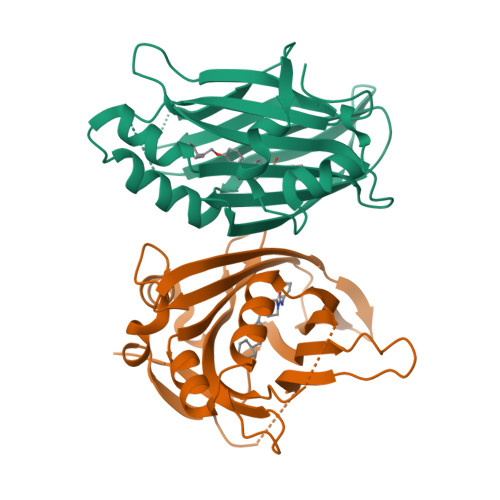
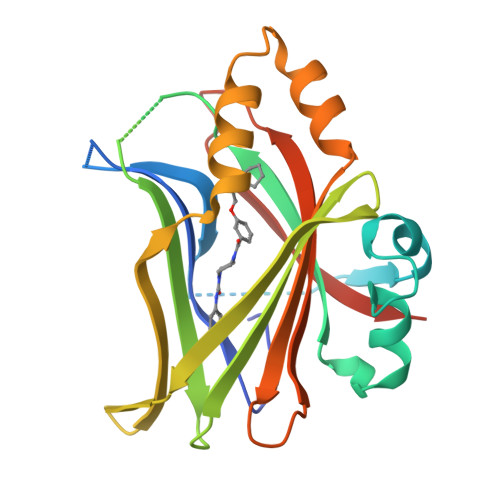
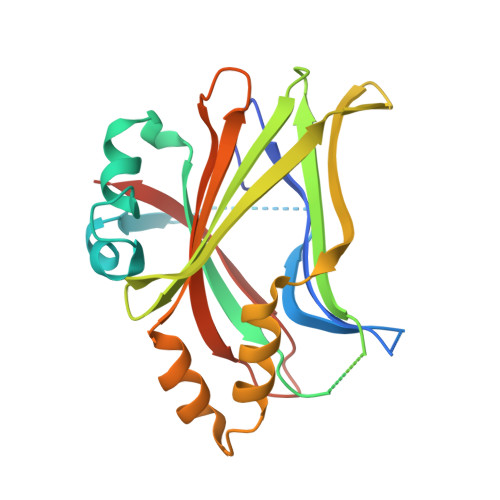
8CUH - PubMed Abstract:
The TEA domain (TEAD) transcription factor forms a transcription co-activation complex with the key downstream effector of the Hippo pathway, YAP/TAZ. TEAD-YAP controls the expression of Hippo-responsive genes involved in cell proliferation, development, and tumorigenesis. Hyperactivation of TEAD-YAP activities is observed in many human cancers and is associated with cancer cell proliferation, survival, and immune evasion. Therefore, targeting the TEAD-YAP complex has emerged as an attractive therapeutic approach. We previously reported that the mammalian TEAD transcription factors (TEAD1-4) possess auto-palmitoylation activities and contain an evolutionarily conserved palmitate-binding pocket (PBP), which allows small-molecule modulation. Since then, several reversible and irreversible inhibitors have been reported by binding to PBP. Here, we report a new class of TEAD inhibitors with a novel binding mode. Representative analog TM2 shows potent inhibition of TEAD auto-palmitoylation both in vitro and in cells. Surprisingly, the co-crystal structure of the human TEAD2 YAP-binding domain (YBD) in complex with TM2 reveals that TM2 adopts an unexpected binding mode by occupying not only the hydrophobic PBP, but also a new side binding pocket formed by hydrophilic residues. RNA-seq analysis shows that TM2 potently and specifically suppresses TEAD-YAP transcriptional activities. Consistently, TM2 exhibits strong antiproliferation effects as a single agent or in combination with a MEK inhibitor in YAP-dependent cancer cells. These findings establish TM2 as a promising small-molecule inhibitor against TEAD-YAP activities and provide new insights for designing novel TEAD inhibitors with enhanced selectivity and potency.
Organizational Affiliation:
Cutaneous Biology Research Center, Massachusetts General Hospital, Harvard Medical School, Charlestown, United States.