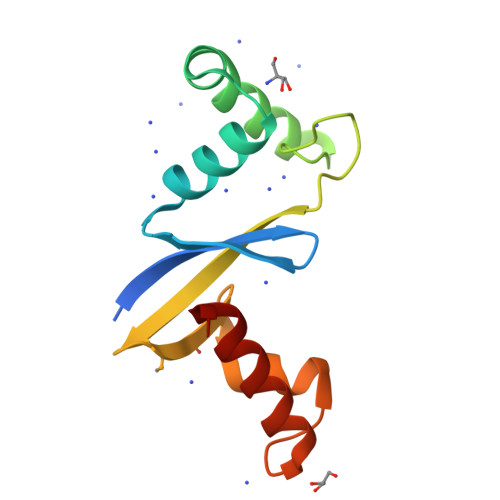
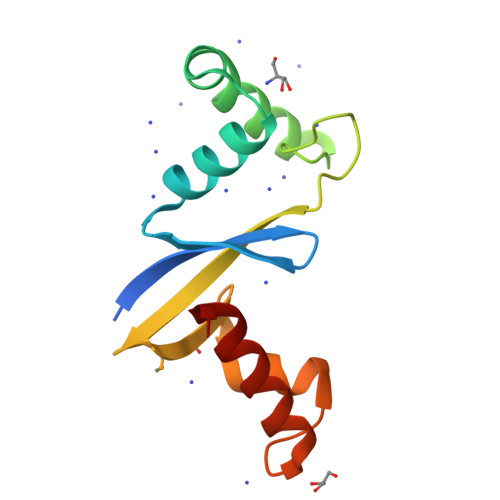
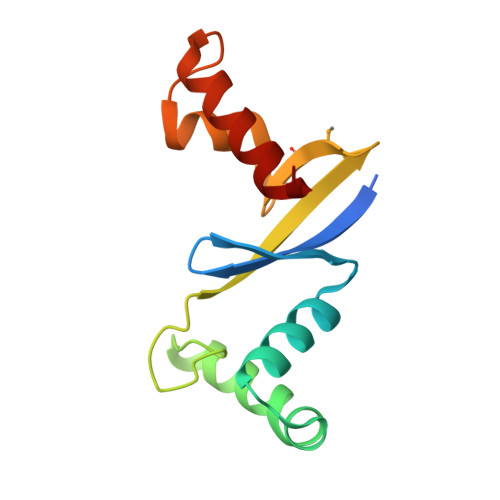
Anti-CRISPR Protein AcrIIC5 Inhibits CRISPR-Cas9 by Occupying the Target DNA Binding Pocket.
Hwang, S., Shah, M., Garcia, B., Hashem, N., Davidson, A.R., Moraes, T.F., Maxwell, K.L.(2023) J Mol Biology 435: 167991-167991
- PubMed: 36736884
- DOI: https://doi.org/10.1016/j.jmb.2023.167991
- Primary Citation of Related Structures:
8F3K - PubMed Abstract:
Anti-CRISPR proteins inhibit CRISPR-Cas immune systems through diverse mechanisms. Previously, the anti-CRISPR protein AcrIIC5 Smu was shown to potently inhibit a type II-C Cas9 from Neisseria meningitidis (Nme1Cas9). In this work, we explore the mechanism of activity of the AcrIIC5 homologue from Neisseria chenwenguii (AcrIIC5 Nch ) and show that it prevents Cas9 binding to target DNA. We show that AcrIIC5 Nch targets the PAM-interacting domain (PID) of Nme1Cas9 for inhibition, agreeing with previous findings for AcrIIC5 Smu , and newly establish that strong binding of the anti-CRISPR requires guide RNA be pre-loaded on Cas9. We determined the crystal structure of AcrIIC5 Nch using X-ray crystallography and identified amino acid residues that are critical for its function. Using a protein docking algorithm we show that AcrIIC5 Nch likely occupies the Cas9 DNA binding pocket, thereby inhibiting target DNA binding through a mechanism similar to that previously described for AcrIIA2 and AcrIIA4.
Organizational Affiliation:
Department of Biochemistry, University of Toronto, 661 University Avenue, Suite 1600, Toronto, Ontario M5G 1M1, Canada. Electronic address: https://twitter.com/s1hwang_21.