Structural Understanding of Fungal Terpene Synthases for the Formation of Linear or Cyclic Terpene Products.
T, R., Sharma, D., Lin, F., Choong, Y.K., Lim, C., Jobichen, C., Zhang, C.(2023) ACS Catal 13: 4949-4959
- PubMed: 37066048
- DOI: https://doi.org/10.1021/acscatal.2c05598
- Primary Citation of Related Structures:
8GY0 - PubMed Abstract:
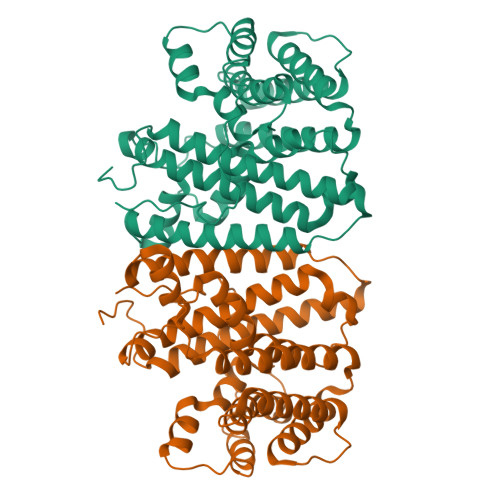
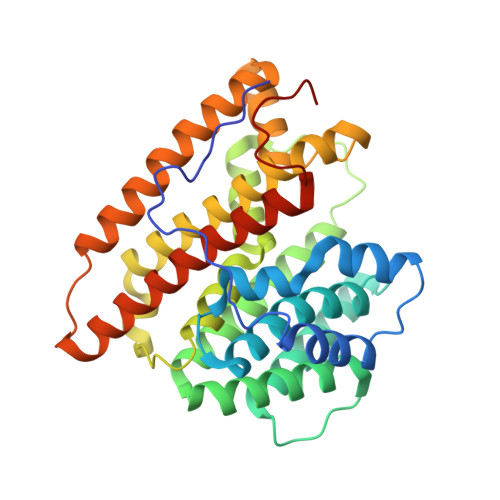
Terpene synthases (TPSs), known gatekeepers of terpenoid diversity, are the main targets for enzyme engineering attempts. To this end, we have determined the crystal structure of Agrocybe pediades linalool synthase (Ap.LS), which has been recently reported to be 44-fold and 287-fold more efficient than bacterial and plant counterparts, respectively. Structure-based molecular modeling followed by in vivo as well as in vitro tests confirmed that the region of 60-69aa and Tyr299 (adjacent to the motif "WxxxxxRY") are essential for maintaining Ap.LS specificity toward a short-chain (C10) acyclic product. Ap.LS Y299 mutants (Y299A, Y299C, Y299G, Y299Q, and Y299S) yielded long-chain (C15) linear or cyclic products. Molecular modeling based on the Ap.LS crystal structure indicated that farnesyl pyrophosphate in the binding pocket of Ap.LS Y299A has less torsion strain energy compared to the wild-type Ap.LS, which can be partially attributed to the larger space in Ap.LS Y299A for better accommodation of the longer chain (C15). Linalool/nerolidol synthase Y298 and humulene synthase Y302 mutations also produced C15 cyclic products similar to Ap.LS Y299 mutants. Beyond the three enzymes, our analysis confirmed that most microbial TPSs have asparagine at the position and produce mainly cyclized products (δ-cadinene, 1,8-cineole, epi-cubebol, germacrene D, β-barbatene, etc.). In contrast, those producing linear products (linalool and nerolidol) typically have a bulky tyrosine. The structural and functional analysis of an exceptionally selective linalool synthase, Ap.LS, presented in this work provides insights into factors that govern chain length (C10 or C15), water incorporation, and cyclization (cyclic vs acyclic) of terpenoid biosynthesis.
Organizational Affiliation:
Singapore Institute of Food and Biotechnology Innovation (SIFBI), Agency for Science, Technology and Research (ASTAR), Singapore 138669, Singapore.