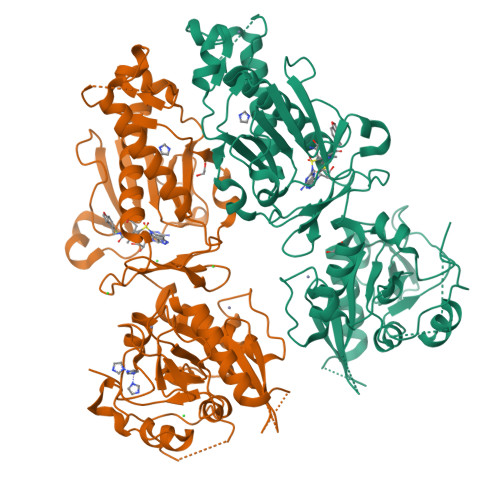
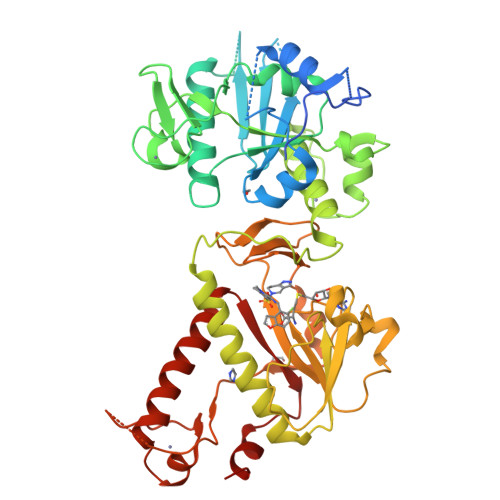
Small-molecule inhibition of SARS-CoV-2 NSP14 RNA cap methyltransferase.
Meyer, C., Garzia, A., Miller, M.W., Huggins, D.J., Myers, R.W., Hoffmann, H.H., Ashbrook, A.W., Jannath, S.Y., Liverton, N., Kargman, S., Zimmerman, M., Nelson, A.M., Sharma, V., Dolgov, E., Cangialosi, J., Penalva-Lopez, S., Alvarez, N., Chang, C.W., Oswal, N., Gonzalez, I., Rasheed, R., Goldgirsh, K., Davis, J.A., Ramos-Espiritu, L., Menezes, M.R., Larson, C., Nitsche, J., Ganichkin, O., Alwaseem, H., Molina, H., Steinbacher, S., Glickman, J.F., Perlin, D.S., Rice, C.M., Meinke, P.T., Tuschl, T.(2025) Nature 637: 1178-1185
- PubMed: 39663451
- DOI: https://doi.org/10.1038/s41586-024-08320-0
- Primary Citation of Related Structures:
8R7B - PubMed Abstract:
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) 1 . The rapid development of highly effective vaccines 2,3 against SARS-CoV-2 has altered the trajectory of the pandemic, and antiviral therapeutics 4 have further reduced the number of COVID-19 hospitalizations and deaths. Coronaviruses are enveloped, positive-sense, single-stranded RNA viruses that encode various structural and non-structural proteins, including those critical for viral RNA replication and evasion from innate immunity 5 . Here we report the discovery and development of a first-in-class non-covalent small-molecule inhibitor of the viral guanine-N7 methyltransferase (MTase) NSP14. High-throughput screening identified RU-0415529, which inhibited SARS-CoV-2 NSP14 by forming a unique ternary S-adenosylhomocysteine (SAH)-bound complex. Hit-to-lead optimization of RU-0415529 resulted in TDI-015051 with a dissociation constant (K d ) of 61 pM and a half-maximal effective concentration (EC 50 ) of 11 nM, inhibiting virus infection in a cell-based system. TDI-015051 also inhibited viral replication in primary small airway epithelial cells and in a transgenic mouse model of SARS CoV-2 infection with an efficacy comparable with the FDA-approved reversible covalent protease inhibitor nirmatrelvir 6 . The inhibition of viral cap methylases as an antiviral strategy is also adaptable to other pandemic viruses.
Organizational Affiliation:
Laboratory for RNA Molecular Biology, The Rockefeller University, New York, NY, USA.