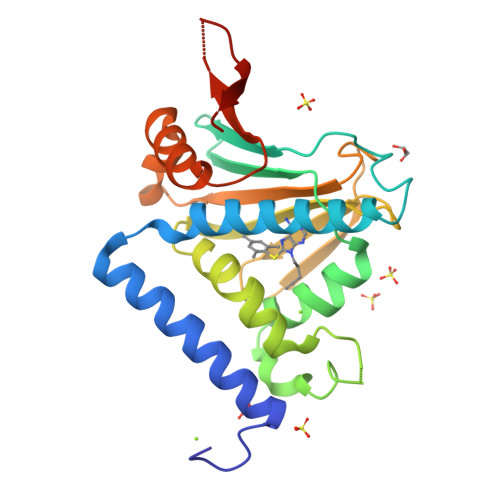
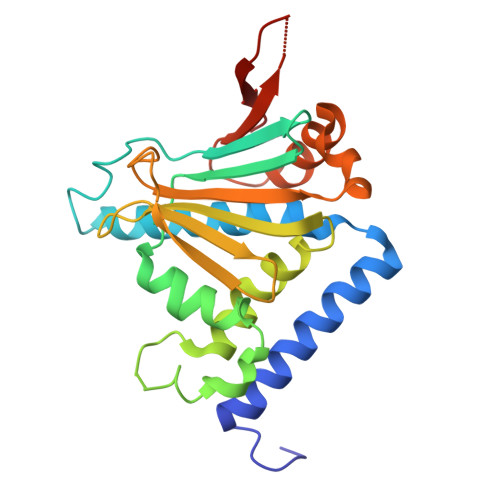
Selective Inhibition of hsp90 Paralogs: Uncovering the Role of Helix 1 in Grp94-Selective Ligand Binding.
Que, N.L.S., Seidler, P.M., Aw, W.J., Chiosis, G., Gewirth, D.T.(2025) Proteins 93: 654-672
- PubMed: 39473058
- DOI: https://doi.org/10.1002/prot.26756
- Primary Citation of Related Structures:
8SBT, 8SSV, 8TF0 - PubMed Abstract:
Grp94 is the endoplasmic reticulum paralog of the hsp90 family of chaperones, which have been targeted for therapeutic intervention via their highly conserved ATP binding sites. The design of paralog-selective inhibitors relies on understanding the protein structural elements that drive higher affinity in selective inhibitors. Here, we determined the structures of Grp94 and Hsp90 in complex with the Grp94-selective inhibitor PU-H36, and of Grp94 with the non-selective inhibitor PU-H71. In Grp94, PU-H36 derives its higher affinity by utilizing Site 2, a Grp94-specific side pocket adjoining the ATP binding cavity, but in Hsp90 PU-H36 occupies Site 1, a side pocket that is accessible in all paralogs with which it makes lower affinity interactions. The structure of Grp94 in complex with PU-H71 shows only Site 1 binding. While changes in the conformation of helices 4 and 5 in the N-terminal domain occur when ligands bind to Site 1 of both Hsp90 and Grp94, large conformational shifts that also involve helix 1 are associated with the engagement of the Site 2 pocket in Grp94 only. Site 2 in Hsp90 is blocked and its helix 1 conformation is insensitive to ligand binding. To understand the role of helix 1 in ligand selectivity, we tested the binding of PU-H36 and other Grp94-selective ligands to chimeric Grp94/Hsp90 constructs. These studies show that helix 1 is the major determinant of selectivity for Site 2 targeted ligands and also influences the rate of ATPase activity in Hsp90 paralogs.
Organizational Affiliation:
Hauptman Woodward Medical Research Institute, Buffalo, New York, USA.