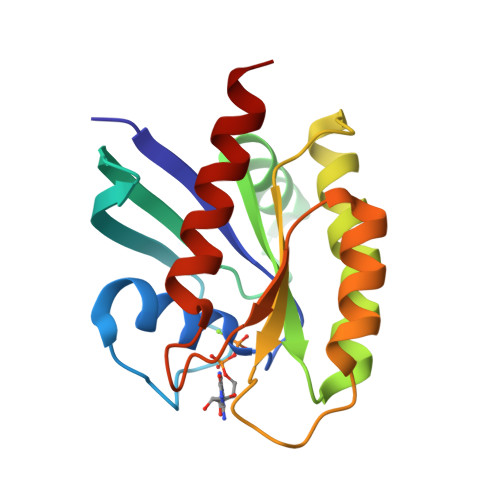
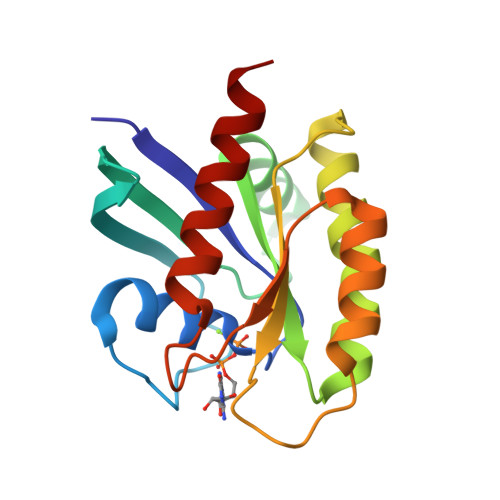
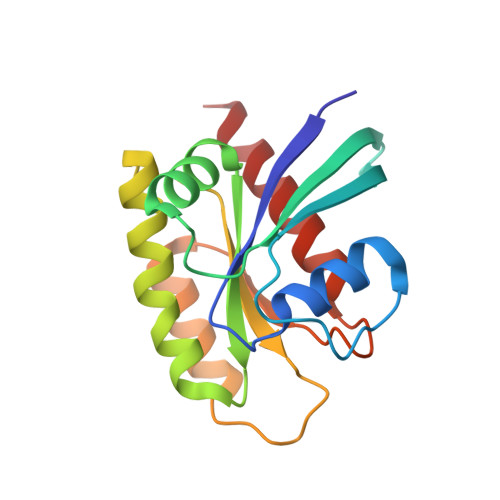
Revealing Functional Hotspots: Temperature-Dependent Crystallography of K-RAS Highlights Allosteric and Druggable Sites.
Deck, S.L., Xu, M., Milano, S.K., Cerione, R.A.(2025) bioRxiv
- PubMed: 40060414
- DOI: https://doi.org/10.1101/2025.02.27.639303
- Primary Citation of Related Structures:
8TVK - PubMed Abstract:
K-RAS mutations drive oncogenesis in multiple cancers, yet the lack of druggable sites has long hindered therapeutic development. Here, we use multi-temperature X-ray crystallography (MT-XRC) to capture functionally relevant K-RAS conformations across a temperature gradient, spanning cryogenic to physiological and even "fever" conditions, and show how cryogenic conditions may obscure key dynamic states as targets for new drug development. This approach revealed a temperature-dependent conformational landscape of K-RAS, shedding light on the dynamic nature of key regions. We identified significant conformational changes occurring at critical sites, including known allosteric and drug-binding pockets, which were hidden under cryogenic conditions but later discovered to be critically important for drug-protein interactions and inhibitor design. These structural changes align with regions previously highlighted by large-scale mutational studies as functionally significant. However, our MT-XRC analysis provides precise structural snapshots, capturing the exact conformations of these potentially important allosteric sites in unprecedented detail. Our findings underscore the necessity of advancing tools like MT-XRC to visualize conformational transitions that may be important in signal propagation which are missed by standard cryogenic XRC and to address hard-to-drug targets through rational drug design. This approach not only provides unique structural insights into K-RAS signaling events and identifies new potential sites to target with drug candidates but also establishes a powerful framework for discovering therapeutic opportunities against other challenging drug targets.
Organizational Affiliation:
Department of Chemistry and Chemical Biology, Cornell University, Ithaca, NY 14853.